The CXCR4–CXCL12-Axis Is of Prognostic Relevance in DLBCL and Its Antagonists Exert Pro-Apoptotic Effects In Vitro
Abstract
:1. Introduction
2. Results
2.1. High Expression of CXCR4 Is Associated with Poor Clinical Outcome in DLBCL
2.2. CXCR4 Is Somatically Unmutated in DLCBL
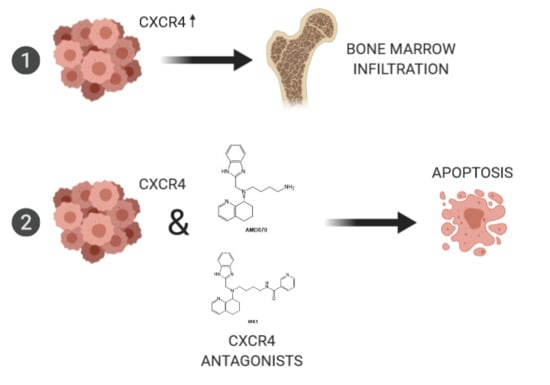
2.3. CXCR4-CXCL12-Axis Is Associated with Bone Marrow Infiltration in DLBCL
2.4. Treatment of Lymphoma Cell Lines with CXCR4 Antagonists Induced Apoptosis
2.5. WK1 and AMD070 Increased Expression of Pro-Apoptotic BCL2-Members
2.6. WK1 Treatment Causes Downregulation of JNK-, ERK1/2, and NF-κB/ BCR-Targets
3. Discussion
4. Materials and Methods
4.1. Patient Samples
4.2. Sequencing of CXCR4
4.3. Cell Lines and Cell Culture
4.4. Synthesis of WK1
4.5. RNA Extraction and RQ-PCR
4.6. Immunohistochemistry
4.7. CXCL12 Binding Assay
4.8. Assessment of Cell Growth
4.9. Apoptosis Assays
4.10. Migration Assay
4.11. Microarray Analysis
4.12. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pasqualucci, L.; Dalla-Favera, R. The genetic landscape of diffuse large B-cell lymphoma. Semin. Hematol. 2015, 52, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Bouska, A.; McKeithan, T.W.; Deffenbacher, K.E.; Lachel, C.; Wright, G.W.; Iqbal, J.; Smith, L.M.; Zhang, W.; Kucuk, C.; Rinaldi, A.; et al. Genome-wide copy-number analyses reveal genomic abnormalities involved in transformation of follicular lymphoma. Blood 2014, 123, 1681–1690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Jong, D.; Balagué Ponz, O. The molecular background of aggressive B cell lymphomas as a basis for targeted therapy. J. Pathol. 2011, 223, 274–282. [Google Scholar] [CrossRef] [PubMed]
- Lenz, G.; Staudt, L.M. Aggressive lymphomas. N. Engl. J. Med. 2010, 362, 1417–1429. [Google Scholar] [CrossRef] [PubMed]
- Hans, C.P.; Weisenburger, D.D.; Greiner, T.C.; Gascoyne, R.D.; Delabie, J.; Ott, G.; Müller-Hermelink, H.K.; Campo, E.; Braziel, R.M.; Jaffe, E.S.; et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood 2004, 103, 275–282. [Google Scholar] [CrossRef] [PubMed]
- D’Apuzzo, M.; Rolink, A.; Loetscher, M.; Hoxie, J.A.; Clark-Lewis, I.; Melchers, F.; Baggiolini, M.; Moser, B. The chemokine SDF-1, stromal cell-derived factor 1, attracts early stage B cell precursors via the chemokine receptor CXCR4. Eur. J. Immunol. 1997, 27, 1788–1793. [Google Scholar] [CrossRef] [PubMed]
- Egawa, T.; Kawabata, K.; Kawamoto, H.; Amada, K.; Okamoto, R.; Fujii, N.; Kishimoto, T.; Katsura, Y.; Nagasawa, T. The earliest stages of B cell development require a chemokine stromal cell-derived factor/pre-B cell growth-stimulating factor. Immunity 2001, 15, 323–334. [Google Scholar] [CrossRef]
- Nie, Y.; Waite, J.; Brewer, F.; Sunshine, M.-J.; Littman, D.R.; Zou, Y.-R. The role of CXCR4 in maintaining peripheral B cell compartments and humoral immunity. J. Exp. Med. 2004, 200, 1145–1156. [Google Scholar] [CrossRef] [PubMed]
- Okada, T.; Ngo, V.N.; Ekland, E.H.; Förster, R.; Lipp, M.; Littman, D.R.; Cyster, J.G. Chemokine requirements for B cell entry to lymph nodes and Peyer’s patches. J. Exp. Med. 2002, 196, 65–75. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Xu-Monette, Z.Y.; Deng, L.; Shen, Q.; Manyam, G.C.; Martinez-Lopez, A.; Zhang, L.; Montes-Moreno, S.; Visco, C.; Tzankov, A.; et al. Dysregulated CXCR4 expression promotes lymphoma cell survival and independently predicts disease progression in germinal center B-cell-like diffuse large B-cell lymphoma. Oncotarget 2015, 6, 5597–5614. [Google Scholar] [CrossRef] [Green Version]
- Deutsch, A.J.A.; Steinbauer, E.; Hofmann, N.A.; Strunk, D.; Gerlza, T.; Beham-Schmid, C.; Schaider, H.; Neumeister, P. Chemokine receptors in gastric MALT lymphoma: loss of CXCR4 and upregulation of CXCR7 is associated with progression to diffuse large B-cell lymphoma. Mod. Pathol. 2013, 26, 182–194. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Zhang, L.; Li, G.; Liu, W.; Tang, W.; Zhang, H.; Luan, J.; Gao, L.; Wang, X. CXCR4 and CCR7 Expression in Primary Nodal Diffuse Large B-Cell Lymphoma-A Clinical and Immunohistochemical Study. Am. J. Med. Sci. 2019, 357, 302–310. [Google Scholar] [CrossRef] [PubMed]
- Laursen, M.B.; Reinholdt, L.; Schönherz, A.A.; Due, H.; Jespersen, D.S.; Grubach, L.; Ettrup, M.S.; Røge, R.; Falgreen, S.; Sørensen, S.; et al. High CXCR4 expression impairs rituximab response and the prognosis of R-CHOP-treated diffuse large B-cell lymphoma patients. Oncotarget 2019, 10, 717–731. [Google Scholar] [CrossRef] [PubMed]
- Moreno, M.J.; Bosch, R.; Dieguez-Gonzalez, R.; Novelli, S.; Mozos, A.; Gallardo, A.; Pavón, M.Á.; Céspedes, M.V.; Grañena, A.; Alcoceba, M.; et al. CXCR4 expression enhances diffuse large B cell lymphoma dissemination and decreases patient survival. J. Pathol. 2015, 235, 445–455. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.C.; Seo, J.; Kang, B.W.; Moon, J.H.; Chae, Y.S.; Lee, S.J.; Lee, Y.J.; Han, S.; Seo, S.K.; Kim, J.G.; et al. Clinical significance of nuclear factor κB and chemokine receptor CXCR4 expression in patients with diffuse large B-cell lymphoma who received rituximab-based therapy. Korean J. Intern. Med. 2014, 29, 785–792. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.-Z.; Shen, J.-K.; Zhao, S.-Q.; Li, J.-M. Clinical significance of chemokine receptor CXCR4 and mammalian target of rapamycin (mTOR) expression in patients with diffuse large B-cell lymphoma. Leuk. Lymphoma 2018, 59, 1451–1460. [Google Scholar] [CrossRef] [PubMed]
- Balabanian, K.; Lagane, B.; Infantino, S.; Chow, K.Y.C.; Harriague, J.; Moepps, B.; Arenzana-Seisdedos, F.; Thelen, M.; Bachelerie, F. The chemokine SDF-1/CXCL12 binds to and signals through the orphan receptor RDC1 in T lymphocytes. J. Biol. Chem. 2005, 280, 35760–35766. [Google Scholar] [CrossRef] [PubMed]
- Debnath, B.; Xu, S.; Grande, F.; Garofalo, A.; Neamati, N. Small molecule inhibitors of CXCR4. Theranostics 2013, 3, 47–75. [Google Scholar] [CrossRef]
- Shakir, M.; Tang, D.; Zeh, H.J.; Tang, S.W.; Anderson, C.J.; Bahary, N.; Lotze, M.T. The chemokine receptors CXCR4/CXCR7 and their primary heterodimeric ligands CXCL12 and CXCL12/high mobility group box 1 in pancreatic cancer growth and development: finding flow. Pancreas 2015, 44, 528–534. [Google Scholar] [CrossRef]
- Lenz, G.; Wright, G.; Dave, S.S.; Xiao, W.; Powell, J.; Zhao, H.; Xu, W.; Tan, B.; Goldschmidt, N.; Iqbal, J.; et al. Stromal gene signatures in large-B-cell lymphomas. N. Engl. J. Med. 2008, 359, 2313–2323. [Google Scholar] [CrossRef]
- Hunter, Z.R.; Xu, L.; Yang, G.; Zhou, Y.; Liu, X.; Cao, Y.; Manning, R.J.; Tripsas, C.; Patterson, C.J.; Sheehy, P.; et al. The genomic landscape of Waldenstrom macroglobulinemia is characterized by highly recurring MYD88 and WHIM-like CXCR4 mutations, and small somatic deletions associated with B-cell lymphomagenesis. Blood 2014, 123, 1637–1646. [Google Scholar] [CrossRef] [PubMed]
- Treon, S.P.; Cao, Y.; Xu, L.; Yang, G.; Liu, X.; Hunter, Z.R. Somatic mutations in MYD88 and CXCR4 are determinants of clinical presentation and overall survival in Waldenstrom macroglobulinemia. Blood 2014, 123, 2791–2796. [Google Scholar] [CrossRef] [PubMed]
- Home-SNP-NCBI. Available online: https://0-www-ncbi-nlm-nih-gov.brum.beds.ac.uk/snp/ (accessed on 23 September 2019).
- Beider, K.; Ribakovsky, E.; Abraham, M.; Wald, H.; Weiss, L.; Rosenberg, E.; Galun, E.; Avigdor, A.; Eizenberg, O.; Peled, A.; et al. Targeting the CD20 and CXCR4 pathways in non-hodgkin lymphoma with rituximab and high-affinity CXCR4 antagonist BKT140. Clin. Cancer Res. 2013, 19, 3495–3507. [Google Scholar] [CrossRef] [PubMed]
- Amini, R.-M.; Berglund, M.; Rosenquist, R.; von Heideman, A.; Lagercrantz, S.; Thunberg, U.; Bergh, J.; Sundström, C.; Glimelius, B.; Enblad, G. A Novel B-cell Line (U-2932) Established from a Patient with Diffuse Large B-cell Lymphoma Following Hodgkin Lymphoma. Leuk. Lymphoma 2002, 43, 2179–2189. [Google Scholar] [CrossRef] [PubMed]
- Klanova, M.; Andera, L.; Brazina, J.; Svadlenka, J.; Benesova, S.; Soukup, J.; Prukova, D.; Vejmelkova, D.; Jaksa, R.; Helman, K.; et al. Targeting of BCL2 Family Proteins with ABT-199 and Homoharringtonine Reveals BCL2- and MCL1-Dependent Subgroups of Diffuse Large B-Cell Lymphoma. Clin. Cancer Res. 2016, 22, 1138–1149. [Google Scholar] [CrossRef] [PubMed]
- Masir, N.; Campbell, L.J.; Jones, M.; Mason, D.Y. Pseudonegative BCL2 protein expression in a t(14;18) translocation positive lymphoma cell line: a need for an alternative BCL2 antibody. Pathology 2010, 42, 212–216. [Google Scholar] [CrossRef] [PubMed]
- Finke, J.; Fritzen, R.; Ternes, P.; Trivedi, P.; Bross, K.J.; Lange, W.; Mertelsmann, R.; Dolken, G. Expression of bcl-2 in Burkitt’s lymphoma cell lines: induction by latent Epstein-Barr virus genes. Blood 1992, 80, 459. [Google Scholar]
- Young, R.M.; Shaffer, A.L.; Phelan, J.D.; Staudt, L.M. B-cell receptor signaling in diffuse large B-cell lymphoma. Semin. Hematol. 2015, 52, 77–85. [Google Scholar] [CrossRef]
- Dai, B.; Zhao, X.F.; Mazan-Mamczarz, K.; Hagner, P.; Corl, S.; Bahassi, E.M.; Lu, S.; Stambrook, P.J.; Shapiro, P.; Gartenhaus, R.B. Functional and molecular interactions between ERK and CHK2 in diffuse large B-cell lymphoma. Nat. Commun. 2011, 2, 402. [Google Scholar] [CrossRef] [Green Version]
- Tian, X.; Pelton, A.; Shahsafaei, A.; Dorfman, D.M. Differential expression of enhancer of zeste homolog 2 (EZH2) protein in small cell and aggressive B-cell non-Hodgkin lymphomas and differential regulation of EZH2 expression by p-ERK1/2 and MYC in aggressive B-cell lymphomas. Mod. Pathol. 2016, 29, 1050–1057. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, T.K.; Jordan, N.; Friedberg, J.; Fisher, R.I.; Dent, P.; Grant, S. Inhibition of MEK/ERK1/2 sensitizes lymphoma cells to sorafenib-induced apoptosis. Leuk. Res. 2010, 34, 379–386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmid, C.A.; Robinson, M.D.; Scheifinger, N.A.; Müller, S.; Cogliatti, S.; Tzankov, A.; Müller, A. DUSP4 deficiency caused by promoter hypermethylation drives JNK signaling and tumor cell survival in diffuse large B cell lymphoma. J. Exp. Med. 2015, 212, 775–792. [Google Scholar] [CrossRef] [PubMed]
- Grill, C.; Gheyas, F.; Dayananth, P.; Jin, W.; Ding, W.; Qiu, P.; Wang, L.; Doll, R.J.; English, J.M. Analysis of the ERK1,2 transcriptome in mammary epithelial cells. Biochem. J. 2004, 381, 635–644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herman, S.E.M.; Mustafa, R.Z.; Gyamfi, J.A.; Pittaluga, S.; Chang, S.; Chang, B.; Farooqui, M.; Wiestner, A. Ibrutinib inhibits BCR and NF-κB signaling and reduces tumor proliferation in tissue-resident cells of patients with CLL. Blood 2014, 123, 3286–3295. [Google Scholar] [CrossRef] [PubMed]
- Stein, J.V.; Nombela-Arrieta, C. Chemokine control of lymphocyte trafficking: a general overview. Immunology 2005, 116, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Cojoc, M.; Peitzsch, C.; Trautmann, F.; Polishchuk, L.; Telegeev, G.D.; Dubrovska, A. Emerging targets in cancer management: role of the CXCL12/CXCR4 axis. OncoTargets Ther. 2013, 6, 1347–1361. [Google Scholar] [CrossRef]
- Sun, X.; Cheng, G.; Hao, M.; Zheng, J.; Zhou, X.; Zhang, J.; Taichman, R.S.; Pienta, K.J.; Wang, J. CXCL12/CXCR4/CXCR7 chemokine axis and cancer progression. Cancer Metastasis Rev. 2010, 29, 709–722. [Google Scholar] [CrossRef] [PubMed]
- Gangadhar, T.; Nandi, S.; Salgia, R. The role of chemokine receptor CXCR4 in lung cancer. Cancer Biol. Ther. 2010, 9, 409–416. [Google Scholar] [CrossRef]
- Weber, T.S. Cell Cycle-Associated CXCR4 Expression in Germinal Center B Cells and Its Implications on Affinity Maturation. Front. Immunol. 2018, 9, 1313. [Google Scholar] [CrossRef] [Green Version]
- Casulo, C.; Burack, W.R.; Friedberg, J.W. Transformed follicular non-Hodgkin lymphoma. Blood 2015, 125, 40–47. [Google Scholar] [CrossRef] [Green Version]
- Moreno, M.J.; Gallardo, A.; Novelli, S.; Mozos, A.; Aragó, M.; Pavón, M.Á.; Céspedes, M.V.; Pallarès, V.; Falgàs, A.; Alcoceba, M.; et al. CXCR7 expression in diffuse large B-cell lymphoma identifies a subgroup of CXCR4+ patients with good prognosis. PLoS ONE 2018, 13, e0198789. [Google Scholar] [CrossRef] [PubMed]
- Döring, Y.; Pawig, L.; Weber, C.; Noels, H. The CXCL12/CXCR4 chemokine ligand/receptor axis in cardiovascular disease. Front. Physiol. 2014, 5, 212. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Gao, L.; Luan, J.; Zhang, H.; Xiao, T. C-X-C Chemokine Receptor 4 in Diffuse Large B Cell Lymphoma: Achievements and Challenges. Acta Haematol. 2019, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Guo, S.; Liu, M.; Burow, M.E.; Wang, G. Targeting CXCL12/CXCR4 Axis in Tumor Immunotherapy. Curr. Med. Chem. 2017. [CrossRef] [PubMed]
- Sehn, L.H.; Donaldson, J.; Chhanabhai, M.; Fitzgerald, C.; Gill, K.; Klasa, R.; MacPherson, N.; O’Reilly, S.; Spinelli, J.J.; Sutherland, J.; et al. Introduction of Combined CHOP Plus Rituximab Therapy Dramatically Improved Outcome of Diffuse Large B-Cell Lymphoma in British Columbia. JCO 2005, 23, 5027–5033. [Google Scholar] [CrossRef] [PubMed]
- Piovan, E.; Tosello, V.; Indraccolo, S.; Masiero, M.; Persano, L.; Esposito, G.; Zamarchi, R.; Ponzoni, M.; Chieco-Bianchi, L.; Dalla-Favera, R.; et al. Differential regulation of hypoxia-induced CXCR4 triggering during B-cell development and lymphomagenesis. Cancer Res. 2007, 67, 8605–8614. [Google Scholar] [CrossRef] [PubMed]
- Oh, Y.S.; Kim, H.Y.; Song, I.-C.; Yun, H.-J.; Jo, D.-Y.; Kim, S.; Lee, H.J. Hypoxia induces CXCR4 expression and biological activity in gastric cancer cells through activation of hypoxia-inducible factor-1α. Oncol. Rep. 2012, 28, 2239–2246. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Cai, C.; Zhao, G.; Qiu, X.; Zhao, H.; Ma, Q.; Tian, L.; Li, X.; Hu, Y.; Liao, B.; et al. Hypoxia promotes migration and induces CXCR4 expression via HIF-1α activation in human osteosarcoma. PLoS ONE 2014, 9, e90518. [Google Scholar] [CrossRef]
- Schioppa, T.; Uranchimeg, B.; Saccani, A.; Biswas, S.K.; Doni, A.; Rapisarda, A.; Bernasconi, S.; Saccani, S.; Nebuloni, M.; Vago, L.; et al. Regulation of the chemokine receptor CXCR4 by hypoxia. J. Exp. Med. 2003, 198, 1391–1402. [Google Scholar] [CrossRef]
- Chatterjee, S.; Behnam Azad, B.; Nimmagadda, S. The intricate role of CXCR4 in cancer. Adv. Cancer Res. 2014, 124, 31–82. [Google Scholar] [CrossRef]
- Hiraga, T. Hypoxic Microenvironment and Metastatic Bone Disease. Int. J. Mol. Sci. 2018, 19, 3523. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Wei, Y.; Xia, J.; Wang, S.; Wu, J.; Chen, F.; Huang, G.; Chen, J. CXCL12-CXCR4 contributes to the implication of bone marrow in cancer metastasis. Future Oncol. 2014, 10, 749–759. [Google Scholar] [CrossRef] [PubMed]
- de Clercq, E. Recent advances on the use of the CXCR4 antagonist plerixafor (AMD3100, Mozobil™) and potential of other CXCR4 antagonists as stem cell mobilizers. Pharmacol. Ther. 2010, 128, 509–518. [Google Scholar] [CrossRef] [PubMed]
- Uchida, D.; Kuribayashi, N.; Kinouchi, M.; Sawatani, Y.; Shimura, M.; Mori, T.; Hasegawa, T.; Miyamoto, Y.; Kawamata, H. Effect of a novel orally bioavailable CXCR4 inhibitor, AMD070, on the metastasis of oral cancer cells. Oncol. Rep. 2018, 40, 303–308. [Google Scholar] [CrossRef] [PubMed]
- Beider, K.; Darash-Yahana, M.; Blaier, O.; Koren-Michowitz, M.; Abraham, M.; Wald, H.; Wald, O.; Galun, E.; Eizenberg, O.; Peled, A.; et al. Combination of imatinib with CXCR4 antagonist BKT140 overcomes the protective effect of stroma and targets CML in vitro and in vivo. Mol. Cancer Ther. 2014, 13, 1155–1169. [Google Scholar] [CrossRef] [PubMed]
- Fahham, D.; Weiss, I.D.; Abraham, M.; Beider, K.; Hanna, W.; Shlomai, Z.; Eizenberg, O.; Zamir, G.; Izhar, U.; Shapira, O.M.; et al. In vitro and in vivo therapeutic efficacy of CXCR4 antagonist BKT140 against human non-small cell lung cancer. J. Thorac. Cardiovasc. Surg. 2012, 144, 1167–1175.e1. [Google Scholar] [CrossRef] [PubMed]
- Swerdlow, S.H.; Campo, E.; Harris, N.L.; Jaffe, E.S.; Pileri, S.A.; Stein, H.; Thiele, J. (Eds.) WHO classification of tumours of haematopoietic and lymphoid tissues, Revised 4th ed.; International Agency for Research on Cancer: Lyon, France, 2017; ISBN 928324494X. [Google Scholar]
- Fechter, K.; Feichtinger, J.; Prochazka, K.; Unterluggauer, J.J.; Pansy, K.; Steinbauer, E.; Pichler, M.; Haybaeck, J.; Prokesch, A.; Greinix, H.T.; et al. Cytoplasmic location of NR4A1 in aggressive lymphomas is associated with a favourable cancer specific survival. Sci. Rep. 2018, 8. [Google Scholar] [CrossRef] [PubMed]
- Davies, A.J.; Rosenwald, A.; Wright, G.; Lee, A.; Last, K.W.; Weisenburger, D.D.; Chan, W.C.; Delabie, J.; Braziel, R.M.; Campo, E.; et al. Transformation of follicular lymphoma to diffuse large B-cell lymphoma proceeds by distinct oncogenic mechanisms. Br. J. Haematol. 2007, 136, 286–293. [Google Scholar] [CrossRef] [PubMed]
- Deutsch, A.J.A.; Rinner, B.; Pichler, M.; Prochazka, K.; Pansy, K.; Bischof, M.; Fechter, K.; Hatzl, S.; Feichtinger, J.; Wenzl, K.; et al. NR4A3 Suppresses Lymphomagenesis through Induction of Proapoptotic Genes. Cancer Res. 2017, 77, 2375–2386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deutsch, A.J.A.; Rinner, B.; Wenzl, K.; Pichler, M.; Troppan, K.; Steinbauer, E.; Schwarzenbacher, D.; Reitter, S.; Feichtinger, J.; Tierling, S.; et al. NR4A1-mediated apoptosis suppresses lymphomagenesis and is associated with a favorable cancer-specific survival in patients with aggressive B-cell lymphomas. Blood 2014, 123, 2367–2377. [Google Scholar] [CrossRef] [PubMed]
- Deutsch, A.J.A.; Aigelsreiter, A.; Staber, P.B.; Beham, A.; Linkesch, W.; Guelly, C.; Brezinschek, R.I.; Fruhwirth, M.; Emberger, W.; Buettner, M.; et al. MALT lymphoma and extranodal diffuse large B-cell lymphoma are targeted by aberrant somatic hypermutation. Blood 2007, 109, 3500–3504. [Google Scholar] [CrossRef] [PubMed]
- German Collection of Microorganisms and Cell Cultures GmbH: Welcome to the Leibniz Institute DSMZ. Available online: https://www.dsmz.de/ (accessed on 23 September 2019).
- Lossos, I.S.; Levy, R. Diffuse large B-cell lymphoma: insights gained from gene expression profiling. Int. J. Hematol. 2003, 77, 321–329. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018; Available online: https://www.R-project.org/ (accessed on 24 September 2019).
- Carvalho, B.S.; Irizarry, R.A. A framework for oligonucleotide microarray preprocessing. Bioinformatics 2010, 26, 2363–2367. [Google Scholar] [CrossRef] [PubMed]
- Therneau, T.M.; Grambsch, P.M. Modeling Survival Data: Extending the Cox Model; Springer: New York, NY, USA, 2000; p. xiii + 350. ISBN 0-387-98784-3. [Google Scholar]
- Kassambara, A.; Kosinski, M. Drawing Survival Curves using ‘ggplot2’. Available online: https://cran.r-project.org/web/packages/survminer/survminer.pdf (accessed on 3 September 2019).



| ID | Type | CXCR4 Exon1 | CXCR4 Exon2 | |
|---|---|---|---|---|
| Al1 | ngcb | WT | WT | |
| Al2 | gcb | transformed | WT | WT |
| Al3 | gcb | transformed | WT | rs2228014 |
| Al4 | gcb | transformed | WT | WT |
| Al5 | ngcb | WT | WT | |
| Al6 | gcb | WT | WT | |
| Al8 | gcb | transformed | WT | WT |
| Al9 | gcb | WT | WT | |
| Al10 | ngcb | WT | WT | |
| Al11 | ngcb | WT | rs2228014 | |
| Al12 | ngcb | WT | WT | |
| Al13 | ngcb | WT | WT | |
| Al14 | gcb | transformed | WT | WT |
| Al16 | gcb | transformed | WT | WT |
| Al17 | gcb | transformed | WT | WT |
| Al18 | gcb | WT | WT | |
| Al19 | gcb | transformed | WT | WT |
| Al20 | gcb | transformed | WT | WT |
| Al21 | ngcb | WT | WT | |
| Al22 | gcb | WT | rs2228014 | |
| Al26 | gcb | transformed | WT | WT |
| Al28 | gcb | transformed | WT | WT |
| Al33 | ngcb | WT | WT | |
| Al34 | gcb | transformed | WT | WT |
| Al71 | ngcb | WT | WT | |
| BL2 | cell line | Burkitt like | WT | WT |
| SuDHl4 | cell line | GCB like | WT | WT |
| RI1 | cell line | NGCB like | WT | WT |
| U2932 | cell line | NGCB like | WT | rs2228014 |
| Clinico-Pathologic Parameters | Patients (n = 71) | Proportion | |
|---|---|---|---|
| Subtype | NGCB | 25 | 35.2% |
| GCB | 46 | 64.8% | |
| Sex | female | 37 | 52.11% |
| male | 34 | 47.9% | |
| Age | ≤60a | 19 | 26.8% |
| >60a | 52 | 73.2% | |
| Stage | 1 | 16 | 20.5% |
| 2 | 18 | 23.5% | |
| 3 | 26 | 34% | |
| 4 | 17 | 22% | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pansy, K.; Feichtinger, J.; Ehall, B.; Uhl, B.; Sedej, M.; Roula, D.; Pursche, B.; Wolf, A.; Zoidl, M.; Steinbauer, E.; et al. The CXCR4–CXCL12-Axis Is of Prognostic Relevance in DLBCL and Its Antagonists Exert Pro-Apoptotic Effects In Vitro. Int. J. Mol. Sci. 2019, 20, 4740. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20194740
Pansy K, Feichtinger J, Ehall B, Uhl B, Sedej M, Roula D, Pursche B, Wolf A, Zoidl M, Steinbauer E, et al. The CXCR4–CXCL12-Axis Is of Prognostic Relevance in DLBCL and Its Antagonists Exert Pro-Apoptotic Effects In Vitro. International Journal of Molecular Sciences. 2019; 20(19):4740. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20194740
Chicago/Turabian StylePansy, Katrin, Julia Feichtinger, Barbara Ehall, Barbara Uhl, Miriam Sedej, David Roula, Beata Pursche, Axel Wolf, Manuel Zoidl, Elisabeth Steinbauer, and et al. 2019. "The CXCR4–CXCL12-Axis Is of Prognostic Relevance in DLBCL and Its Antagonists Exert Pro-Apoptotic Effects In Vitro" International Journal of Molecular Sciences 20, no. 19: 4740. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20194740