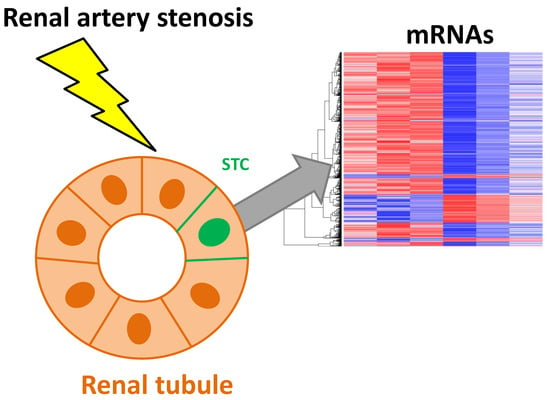
Renal Artery Stenosis Alters Gene Expression in Swine Scattered Tubular-Like Cells
Abstract
:1. Introduction
2. Results
2.1. Systemic Characteristics and Renal Function
2.2. STC Characterization
2.3. mRNAseq Analysis
2.4. Genes Upregulated in RAS-STCs
2.5. Genes Downregulated in RAS-STCs
2.6. Validation of mRNAseq Analysis
2.7. STC Activation and Proliferation
3. Discussion
4. Materials and Methods
4.1. Experimental Design
4.2. STC Isolation and Characterization
4.3. mRNAseq and Bioinformatic Analysis
4.4. Validation of mRNAseq Analysis
4.5. STCs Activation
4.6. STC Proliferation
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviation
| AKI | Acute kidney injury |
| STCs | Scattered tubular-like cells |
| RAS | Renal artery stenosis |
| RBF | Renal blood flow |
| mRNAseq | Messenger RNA sequencing |
| MDCT | Multi-detector computed tomography |
| GFR | Glomerular filtration rate |
| CPM | Counts per million |
| qPCR | Quantitative-polymerase chain reaction |
| BCAM | Linear dichroism basal cell adhesion molecule |
| CADH3 | Cadherin 3 |
| CADH16 | Cadherin 6 |
| EpCAM | Epithelial cellular adhesion molecule |
| COL6A2 | Collagen 6A2 |
| COL18A1 | Collagen 18A1 |
| LAMA5 | Laminin A5 |
| LAMB3 | Laminin B3 |
| POU3F3 | POU Class 3 Homeobox 3 |
| SOX | Sex determining region Y-box |
| PANTHER | Protein Analysis Through Evolutionary Relationships |
| DAVID | Database for Annotation, Visualization, and Integrated Discovery |
| STRING | Search Tool for the Retrieval of Interacting Genes |
| PHA-E | Phaseolus vulgaris erythroagglutinin |
References
- Bussolati, B.; Camussi, G. Therapeutic use of human renal progenitor cells for kidney regeneration. Nat. Rev. Nephrol. 2015, 11, 695–706. [Google Scholar] [CrossRef] [PubMed]
- Kusaba, T.; Lalli, M.; Kramann, R.; Kobayashi, A.; Humphreys, B.D. Differentiated kidney epithelial cells repair injured proximal tubule. Proc. Natl. Acad. Sci. USA 2014, 111, 1527–1532. [Google Scholar] [CrossRef] [PubMed]
- Ronconi, E.; Sagrinati, C.; Angelotti, M.L.; Lazzeri, E.; Mazzinghi, B.; Ballerini, L.; Parente, E.; Becherucci, F.; Gacci, M.; Carini, M.; et al. Regeneration of glomerular podocytes by human renal progenitors. J. Am. Soc. Nephrol. 2009, 20, 322–332. [Google Scholar] [CrossRef] [PubMed]
- Angelotti, M.L.; Ronconi, E.; Ballerini, L.; Peired, A.; Mazzinghi, B.; Sagrinati, C.; Parente, E.; Gacci, M.; Carini, M.; Rotondi, M.; et al. Characterization of renal progenitors committed toward tubular lineage and their regenerative potential in renal tubular injury. Stem Cells 2012, 30, 1714–1725. [Google Scholar] [CrossRef] [PubMed]
- Sagrinati, C.; Netti, G.S.; Mazzinghi, B.; Lazzeri, E.; Liotta, F.; Frosali, F.; Carini, M.; Ronconi, E.; Meini, C.; Gacci, M.; et al. Isolation and characterization of multipotent progenitor cells from the Bowman’s capsule of adult human kidneys. J. Am. Soc. Nephrol. 2006, 17, 2443–2456. [Google Scholar] [CrossRef]
- Grange, C.; Moggio, A.; Tapparo, M.; Porta, S.; Camussi, G.; Bussolati, B. Protective effect and localization by optical imaging of human renal CD133+ progenitor cells in an acute kidney injury model. Physiol. Rep. 2014, 2, e12009. [Google Scholar] [CrossRef]
- Zou, X.; Kwon, S.H.; Jiang, K.; Ferguson, C.M.; Puranik, A.S.; Zhu, X.; Lerman, L.O. Renal scattered tubular-like cells confer protective effects in the stenotic murine kidney mediated by release of extracellular vesicles. Sci. Rep. 2018, 8, 1263. [Google Scholar] [CrossRef]
- Zoccali, C.; Mallamaci, F.; Finocchiaro, P. Atherosclerotic renal artery stenosis: Epidemiology, cardiovascular outcomes, and clinical prediction rules. J. Am. Soc. Nephrol. 2002, 13, S179–S183. [Google Scholar] [CrossRef]
- Textor, S.C.; Lerman, L.O. Paradigm shifts in atherosclerotic renovascular disease: Where are we now? J. Am. Soc. Nephrol. 2015, 26, 2074–2080. [Google Scholar] [CrossRef]
- Lerman, L.O.; Textor, S.C.; Grande, J.P. Mechanisms of tissue injury in renal artery stenosis: Ischemia and beyond. Prog. Cardiovasc. Dis. 2009, 52, 196–203. [Google Scholar] [CrossRef]
- Eirin, A.; Lerman, L.O. Darkness at the end of the tunnel: Poststenotic kidney injury. Physiology 2013, 28, 245–253. [Google Scholar] [CrossRef] [PubMed]
- Aghajani Nargesi, A.; Zhu, X.Y.; Conley, S.M.; Woollard, J.R.; Saadiq, I.M.; Lerman, L.O.; Eirin, A. Renovascular disease induces mitochondrial damage in swine scattered tubular cells. Am. J. Physiol. Renal Physiol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Eirin, A.; Gloviczki, M.L.; Tang, H.; Rule, A.D.; Woollard, J.R.; Lerman, A.; Lerman, L.O.; Textor, S.C. Chronic renovascular hypertension is associated with elevated levels of neutrophil gelatinase-associated lipocalin. Nephrol. Dial. Transplant. 2012, 27, 4153–4161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eirin, A.; Textor, S.C.; Lerman, L.O. Emerging Paradigms in Chronic Kidney Ischemia. Hypertension 2018, 72, 1023–1030. [Google Scholar] [CrossRef] [PubMed]
- Hansson, J.; Hultenby, K.; Cramnert, C.; Pontén, F.; Jansson, H.; Lindgren, D.; Johansson, M.E.; Axelson, H. Evidence for a morphologically distinct and functionally robust cell type in the proximal tubules of human kidney. Hum. Pathol. 2014, 45, 382–393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shirasawa, T.; Akashi, T.; Sakamoto, K.; Takahashi, H.; Maruyama, N.; Hirokawa, K. Gene expression of CD24 core peptide molecule in developing brain and developing non-neural tissues. Dev. Dyn. 1993, 198, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Kemper, K.; Sprick, M.R.; de Bree, M.; Scopelliti, A.; Vermeulen, L.; Hoek, M.; Medema, J.P.; Zeilstra, J.; Pals, S.T.; Mehmet, H.; et al. The AC133 epitope, but not the CD133 protein, is lost upon cancer stem cell differentiation. Cancer Res. 2010, 70, 719–729. [Google Scholar] [CrossRef]
- Metsuyanim, S.; Harari-Steinberg, O.; Buzhor, E.; Omer, D.; Pode-Shakked, N.; Ben-Hur, H.; Dekel, B.; Halperin, R.; Schneider, D. Expression of stem cell markers in the human fetal kidney. PLoS ONE 2009, 4, e6709. [Google Scholar] [CrossRef]
- Akcay, A.; Nguyen, Q.; Edelstein, C.L. Mediators of inflammation in acute kidney injury. Mediat. Inflamm. 2009, 2009, 137072. [Google Scholar] [CrossRef]
- Kumar, S.; Liu, J.; Pang, P.; Krautzberger, A.M.; Reginensi, A.; Akiyama, H.; McMahon, A.P.; Schedl, A.; Humphreys, B.D. Sox9 activation highlights a cellular pathway of renal repair in the acutely injured mammalian Kidney. Cell Rep. 2015, 12, 1325–1338. [Google Scholar] [CrossRef]
- Kang, H.M.; Huang, S.; Reidy, K.; Han, S.H.; Chinga, F.; Susztak, K. Sox9-positive progenitor cells play a key role in renal tubule epithelial regeneration in mice. Cell Rep. 2016, 14, 861–871. [Google Scholar] [CrossRef] [PubMed]
- Barrington, C.; Pezic, D.; Hadjur, S. Chromosome structure dynamics during the cell cycle: A structure to fit every phase. EMBO J. 2017, 36, 2661–2663. [Google Scholar] [CrossRef] [PubMed]
- McGlynn, P. Helicases at the replication fork. Adv. Exp. Med. Biol. 2013, 767, 97–121. [Google Scholar] [CrossRef] [PubMed]
- Witzgall, R.; Brown, D.; Schwarz, C.; Bonventre, J.V. Localization of proliferating cell nuclear antigen, vimentin, c-Fos, and clusterin in the postischemic kidney. Evidence for a heterogenous genetic response among nephron segments, and a large pool of mitotically active and dedifferentiated cells. J. Clin. Investig. 1994, 93, 2175–2188. [Google Scholar] [CrossRef]
- Goldman, R.D.; Khuon, S.; Chou, Y.H.; Opal, P.; Steinert, P.M. The function of intermediate filaments in cell shape and cytoskeletal integrity. J. Cell Biol. 1996, 134, 971–983. [Google Scholar] [CrossRef]
- Forth, S.; Kapoor, T.M. The mechanics of microtubule networks in cell division. J. Cell Biol. 2017, 216, 1525–1531. [Google Scholar] [CrossRef] [Green Version]
- Hinchcliffe, E.H.; Miller, F.J.; Cham, M.; Khodjakov, A.; Sluder, G. Requirement of a centrosomal activity for cell cycle progression through G1 into S phase. Science 2001, 291, 1547–1550. [Google Scholar] [CrossRef]
- Shen, S.Q.; Yan, X.W.; Li, P.T.; Ji, X.H. Analysis of differential gene expression by RNA-seq data in ABCG1 knockout mice. Gene 2019, 689, 24–33. [Google Scholar] [CrossRef]
- Shu-Shan, Z.; Jian-Jun, D.; Cai-Feng, W.; Ting-Yu, Z.; De-Fu, Z. Comparative proteomic analysis of hearts of adult SCNT Bama miniature pigs (Sus scrofa). Theriogenology 2014, 81, 901–905. [Google Scholar] [CrossRef]
- Arce, C.; Lucena, C.; Moreno, A.; Garrido, J.J. Proteomic analysis of intestinal mucosa responses to Salmonella enterica serovar typhimurium in naturally infected pig. Comp. Immunol. Microbiol. Infect. Dis. 2014, 37, 59–67. [Google Scholar] [CrossRef]
- Lerman, L.O.; Schwartz, R.S.; Grande, J.P.; Sheedy, P.F.; Romero, J.C. Noninvasive evaluation of a novel swine model of renal artery stenosis. J. Am. Soc. Nephrol. 1999, 10, 1455–1465. [Google Scholar] [PubMed]
- Krier, J.D.; Ritman, E.L.; Bajzer, Z.; Romero, J.C.; Lerman, A.; Lerman, L.O. Noninvasive measurement of concurrent single-kidney perfusion, glomerular filtration, and tubular function. Am. J. Physiol. Renal Physiol. 2001, 281, F630–F638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eirin, A.; Ebrahimi, B.; Zhang, X.; Zhu, X.Y.; Tang, H.; Crane, J.A.; Lerman, L.O.; Lerman, A.; Textor, S.C. Changes in glomerular filtration rate after renal revascularization correlate with microvascular hemodynamics and inflammation in Swine renal artery stenosis. Circ. Cardiovasc. Interv. 2012, 5, 720–728. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Jia, Y.; Li, C.; Cheng, Q.; Yue, W.; Pei, X. Hyperglycemic Stress Impairs the Stemness Capacity of Kidney Stem Cells in Rats. PLoS ONE 2015, 10, e0139607. [Google Scholar] [CrossRef]
- Zhu, X.Y.; Urbieta-Caceres, V.; Krier, J.D.; Textor, S.C.; Lerman, A.; Lerman, L.O. Mesenchymal stem cells and endothelial progenitor cells decrease renal injury in experimental swine renal artery stenosis through different mechanisms. Stem Cells 2013, 31, 117–125. [Google Scholar] [CrossRef]
- Dudakovic, A.; Camilleri, E.; Riester, S.M.; Lewallen, E.A.; Kvasha, S.; Chen, X.; Krych, A.J.; Radel, D.J.; Anderson, J.M.; Nair, A.A.; et al. High-resolution molecular validation of self-renewal and spontaneous differentiation in adipose-tissue derived human mesenchymal stem cells cultured in human platelet lysate. J. Cell. Biochem. 2014, 115, 1816–1828. [Google Scholar] [CrossRef]
- Kalari, K.R.; Nair, A.A.; Bhavsar, J.D.; O’Brien, D.R.; Davila, J.I.; Bockol, M.A.; Middha, S.; Nie, J.; Tang, X.; Baheti, S.; et al. MAP-RSeq: Mayo Analysis Pipeline for RNA sequencing. BMC Bioinform. 2014, 15, 224. [Google Scholar] [CrossRef]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Gingeras, T.R.; Batut, P.; Chaisson, M. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Wang, L.; Wang, S.; Li, W. RSeQC: Quality control of RNA-seq experiments. Bioinformatics 2012, 28, 2184–2185. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wu, P.; Zhou, J.; Johnson-Pais, T.L.; Lai, Z.; Chowdhury, W.H.; Chen, Y.; Rodriguez, R. XBSeq2: A fast and accurate quantification of differential expression and differential polyadenylation. BMC Bioinform. 2017, 18, 384. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, D.J.; Chen, Y.; Smyth, G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012, 40, 4288–4297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mi, H.; Lazareva-Ulitsky, B.; Loo, R.; Kejariwal, A.; Vandergriff, J.; Rabkin, S.; Kitano, H.; Guo, N.; Muruganujan, A.; Doremieux, O.; et al. The PANTHER database of protein families, subfamilies, functions and pathways. Nucleic Acids Res. 2005, 33, D284–D288. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef]
- Park, E.; Cho, M.; Ki, C.S. Correct use of repeated measures analysis of variance. Korean J. Lab. Med. 2009, 29, 1–9. [Google Scholar] [CrossRef]








| Parameter | Normal | RAS |
|---|---|---|
| Body Weight (Kg) | 50.3 ± 0.6 | 54.3 ± 5.5 |
| Degree of stenosis (%) | 0 | 84.7 ± 12.7 * |
| Systolic blood pressure (mmHg) | 96.3 ± 5.1 | 151.7 ± 33.3 * |
| Diastolic blood pressure (mmHg) | 70.0 ± 6.0 | 118.7 ± 21.7 * |
| Mean arterial pressure (mmHg) | 78.8 ± 5.3 | 129.7 ± 25.5 * |
| Serum creatinine (mg/dL) | 1.2 ± 0.1 | 1.8 ± 0.3 * |
| Cortical volume (mL) | 104.8 ± 4.4 | 58.7 ± 27.1 * |
| Medullary volume (mL) | 19.6 ± 1.0 | 19.5 ± 2.3 |
| Cortical perfusion (mL/min/mL tissue) | 5.4 ± 0.4 | 2.4 ± 0.5 * |
| Medullary perfusion (mL/min/mL tissue) | 2.9 ± 0.3 | 2.7 ± 0.3 |
| RBF (mL/min) | 607.4 ± 42.4 | 315.5 ± 94.4 * |
| GFR (mL/min) | 98.1 ± 9.5 | 52.2 ± 8.3 * |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aghajani Nargesi, A.; Zhu, X.-Y.; Liu, Y.; Tang, H.; Jordan, K.L.; Lerman, L.O.; Eirin, A. Renal Artery Stenosis Alters Gene Expression in Swine Scattered Tubular-Like Cells. Int. J. Mol. Sci. 2019, 20, 5069. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20205069
Aghajani Nargesi A, Zhu X-Y, Liu Y, Tang H, Jordan KL, Lerman LO, Eirin A. Renal Artery Stenosis Alters Gene Expression in Swine Scattered Tubular-Like Cells. International Journal of Molecular Sciences. 2019; 20(20):5069. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20205069
Chicago/Turabian StyleAghajani Nargesi, Arash, Xiang-Yang Zhu, Yuanhang Liu, Hui Tang, Kyra L. Jordan, Lilach O. Lerman, and Alfonso Eirin. 2019. "Renal Artery Stenosis Alters Gene Expression in Swine Scattered Tubular-Like Cells" International Journal of Molecular Sciences 20, no. 20: 5069. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20205069