Chip-Based Digital PCR Approach Provides A Sensitive and Cost-Effective Single-Day Screening Tool for Common Fetal Aneuploidies—A Proof of Concept Study
Abstract
:1. Introduction
2. Results
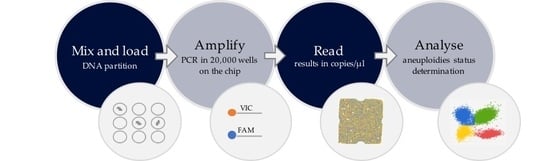
2.1. Workflow of the Chip-Based Digital PCR Experiment
2.2. Validation of the Duplex dPCR Reactions
2.3. Determination of the Cut-Off Value for Aneuploidies
2.4. Validation of Clinical Sample Detection Using Chip-Based dPCR
2.5. dPCR Sensitivity to Mosaicism and Maternal Cell Contamination
3. Discussion
4. Materials and Methods
4.1. Patient Enrollment and Sample Collection
4.2. DNA Extraction
4.3. Chip-Based Digital PCR Workflow
4.4. Statistical Analysis
5. Patents
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| NT | fetal nuchal translucency |
| β-hCG | free beta-human chorionic gonadotropin |
| PAPP-A | pregnancy-associated plasma protein-A |
| FISH | linear dichroism |
| QF-PCR | quantitative fluorescence PCR |
| MLPA | multiplex ligation-dependent probe amplification |
| dPCR | digital PCR |
| CVS | chorionic villus samples |
| ROC | receiver operating characteristic |
| PBS | phosphate buffered saline |
Appendix A
| Gene | Chromosome Location | Primer and Probe Sequences (5′–3′) | Product Size (bp) |
|---|---|---|---|
| MBNL2 | 13q32.1 | F: CTCACCTATCCACAATGCAA | 81 |
| R: GGGATTCAAGCGAATTAACA | |||
| Probe: AGGTGCATCATGGGAACGGC-VIC | |||
| EHZF | 18q11.2 | F: CCAGCTGGTACTTGGAAGAG | 87 |
| R: TGTCGTATGTGGAGCCAAC | |||
| Probe: TCAGTGCCTGCCTGGTTCCC-FAM | |||
| PRDM15 | 21q22.3 | F: ATGTTTCGCCAACTTCTGAG | 89 |
| R: AGAGCTATGGCACAAACCTG | |||
| Probe: TCCCAAACTCTCCTGCCCTGA-FAM | |||
| Noncoding | Xp22.3 | F: GATGAGGAAGGCAATGATCC | 86 |
| R: TTGGCTTTTACCAAATAGGG | |||
| Probe: TGTTTCTCTCTGCCTGCACTGG-VIC | |||
| SRY | Yp11.3 | F: CGCTTAACATAGCAGAAGCA | 84 |
| R: AGTTTCGAACTCTGGCACCT | |||
| Probe: TGTCGCACTCTCCTTGTTTTTGACA-FAM |
References
- Carlson, M.; Vora, N.L. Prenatal Diagnosis: Screening and Diagnostic Tools. Obstet. Gynecol. Clin. North. Am. 2017, 44, 245–256. [Google Scholar] [CrossRef] [PubMed]
- Kagan, K.O.; Wright, D.; Spencer, K.; Molina, F.S.; Nicolaides, K.H. First-trimester screening for trisomy 21 by free beta-human chorionic gonadotropin and pregnancy-associated plasma protein-A: Impact of maternal and pregnancy characteristics. Ultrasound Obs. Gynecol. 2008, 31, 493–502. [Google Scholar] [CrossRef] [PubMed]
- American College of Obstetricians and Gynecologists. ACOG Practice Bulletin No. 88, December 2007. Invasive prenatal testing for aneuploidy. Obstet. Gynecol. 2007, 110, 1459–1467. [Google Scholar]
- Driscoll, D.A.; Gross, S. Clinical practice. Prenatal screening for aneuploidy. N. Engl. J. Med. 2009, 360, 2556–2562. [Google Scholar] [CrossRef] [PubMed]
- EUROCAT Prevalence Tables. Available online: http://www.eurocat-network.eu/accessprevalencedata/prevalencetables (accessed on 29 August 2019).
- Faas, B.H.; Cirigliano, V.; Bui, T.H. Rapid methods for targeted prenatal diagnosis of common chromosome aneuploidies. Semin. Fetal Neonatal Med. 2011, 16, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Nicolini, U.; Lalatta, F.; Natacci, F.; Curcio, C.; Bui, T.H. The introduction of QF-PCR in prenatal diagnosis of fetal aneuploidies: Time for reconsideration. Hum. Reprod. Update. 2004, 10, 541–548. [Google Scholar] [CrossRef] [PubMed]
- Lou, J.; Sun, M.; Zhao, Y.; Ji, Z.; Liu, F.; Li, D.; Xu, W.; Lin, Y.; Liu, Y. Rapid and simultaneous detection of common aneuploidies by quadruplex real-time polymerase chain reaction combined with melting curve analysis. PLoS ONE 2017, 12, e0171886. [Google Scholar] [CrossRef] [PubMed]
- Whale, A.S.; Huggett, J.F.; Cowen, S.; Speirs, V.; Shaw, J.; Ellison, S.; Foy, C.A.; Scott, D.J. Comparison of microfluidic digital PCR and conventional quantitative PCR for measuring copy number variation. Nucleic Acids Res. 2012, 40, e82. [Google Scholar] [CrossRef] [PubMed]
- Vogelstein, B.; Kinzler, K.W. Digital PCR. Proc. Natl. Acad. Sci. USA 1999, 96, 9236–9241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, L.; Meng, Y.; Sui, Z.; Wang, J.; Wu, L.; Fu, B. Comparison of four digital PCR platforms for accurate quantification of DNA copy number of a certified plasmid DNA reference material. Sci Rep. 2015, 5, 13174. [Google Scholar] [CrossRef] [PubMed]
- Huggett, J.F.; Foy, C.A.; Benes, V.; Emslie, K.; Garson, J.A.; Haynes, R.; Hellemans, J.; Kubista, M.; Mueller, R.D.; Nolan, T.; et al. The digital MIQE guidelines: Minimum Information for Publication of Quantitative Digital PCR Experiments. Clin. Chem. 2013, 59, 892–902. [Google Scholar] [CrossRef] [PubMed]
- Basu, A.S. Digital Assays Part I: Partitioning Statistics and Digital PCR. SLAS Technol. 2017, 22, 369–386. [Google Scholar] [CrossRef] [Green Version]
- Quan, P.L.; Sauzade, M.; Brouzes, E. dPCR: A Technology Review. Sensors 2018, 18, 1271. [Google Scholar] [CrossRef] [PubMed]
- Hindson, B.J.; Ness, K.D.; Masquelier, D.A.; Belgrader, P.; Heredia, N.J.; Makarewicz, A.J. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal. Chem. 2011, 83, 8604–8610. [Google Scholar] [CrossRef] [PubMed]
- Sanders, R.; Huggett, J.F.; Bushell, C.A.; Cowen, S.; Scott, D.J.; Foy, C.A. Evaluation of Digital PCR for Absolute DNA Quantification. Anal. Chem. 2011, 83, 6474–6484. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.C.; Blumenfeld, Y.J.; El-Sayed, Y.Y.; Chueh, J.; Quake, S.R. Microfluidic digital PCR enables rapid prenatal diagnosis of fetal aneuploidy. Am. J. Obstet Gynecol. 2009, 200, 543.e1–543.e7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, H.C.; Quake, S.R. Detection of aneuploidy with digital polymerase chain reaction. Anal. Chem. 2007, 79, 7576–7579. [Google Scholar] [CrossRef] [PubMed]






| Chr | Ratio of Slopes in Euploid Samples | Ratio of Slopes in Aneuploid Samples | ||||
|---|---|---|---|---|---|---|
| Mean Ratio | Minimum Ratio | Maximum Ratio | Mean Ratio | Minimum Ratio | Maximum Ratio | |
| 13 | 1.012 | 0.935 | 1.145 | 1.507 | 1.392 | 1.508 |
| 18 | 0.950 | 0.915 | 0.980 | 1.424 | 1.382 | 1.463 |
| 21 | 0.981 | 0.955 | 1.008 | 1.505 | 1.429 | 1.533 |
| Indications | No. of Amniotic Fluid Samples (N = 106) 2 | No. of Chorionic Villus Samples (N = 27) 2 |
|---|---|---|
| Maternal age 1 (range) | 34 ± 5.4 (20–47) | |
| ≤35 years | 57 | 10 |
| >35 years | 49 | 17 |
| Gestational age 1 (range) | 16 ± 3.8 (11–33) | |
| 11–14 weeks | 10 | 25 |
| 15–20 weeks | 86 | 2 |
| >20 weeks | 10 | 0 |
| Clinical characteristics | ||
| Abnormal results of the first trimester screening 2 | 62 | 8 |
| Abnormal results of ultrasound 2 | 45 | 20 |
| Other indications | 6 | 2 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nykel, A.; Kaszkowiak, M.; Fendler, W.; Gach, A. Chip-Based Digital PCR Approach Provides A Sensitive and Cost-Effective Single-Day Screening Tool for Common Fetal Aneuploidies—A Proof of Concept Study. Int. J. Mol. Sci. 2019, 20, 5486. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20215486
Nykel A, Kaszkowiak M, Fendler W, Gach A. Chip-Based Digital PCR Approach Provides A Sensitive and Cost-Effective Single-Day Screening Tool for Common Fetal Aneuploidies—A Proof of Concept Study. International Journal of Molecular Sciences. 2019; 20(21):5486. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20215486
Chicago/Turabian StyleNykel, Anna, Marcin Kaszkowiak, Wojciech Fendler, and Agnieszka Gach. 2019. "Chip-Based Digital PCR Approach Provides A Sensitive and Cost-Effective Single-Day Screening Tool for Common Fetal Aneuploidies—A Proof of Concept Study" International Journal of Molecular Sciences 20, no. 21: 5486. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20215486