Length and Energy Dependence of Low-Energy Electron-Induced Strand Breaks in Poly(A) DNA
Abstract
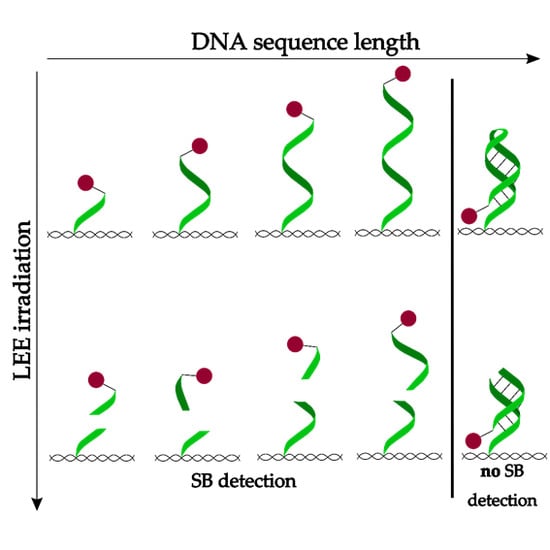
:1. Introduction
2. Results
2.1. Electron Energy Dependence of Strand Break Cross Sections
2.2. Oligonucleotide Length Dependence of Strand Break Cross Sections
3. Materials and Methods
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Sevilla, M.D.; Becker, D.; Kumar, A.; Adhikary, A. Gamma and Ion-Beam Irradiation of DNA: Free Radical Mechanisms, Electron Effects, and Radiation Chemical Track Structure. Radiat. Phys. Chem. 2016, 128, 60–74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boudaïffa, B.; Cloutier, P.; Hunting, D.; Huels, M.A.; Sanche, L. Resonant formation of DNA strand breaks by low-energy (3 to 20 eV) electrons. Science 2000, 287, 1658–1660. [Google Scholar] [PubMed]
- Kumar, A.; Becker, D.; Adhikary, A.; Sevilla, M.D. Reaction of Electrons with DNA: Radiation Damage to Radiosensitization. Int. J. Mol. Sci. 2019, 20, 3998. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Illenberger, E.; Momigny, J. Gaseous Molecular Ions, An Introduction to Elementary Processes Induced by Ionization; aus der Reihe: Topics in Physical Chemistry; Steinkopff Verlag, Springer-Verlag: Darmstadt, Germany, 1992; Volume 2, ISBN 3-7985-0870-4. [Google Scholar]
- Bald, I.; Langer, J.; Tegeder, P.; Ingólfsson, O. From isolated molecules through clusters and condensates to the building blocks of life. Int. J. Mass Spectrom. 2008, 277, 4–25. [Google Scholar] [CrossRef]
- Caldecott, K.W. DNA single-strand break repair. Exp. Cell Res. 2014, 329, 2–8. [Google Scholar] [CrossRef]
- Pandey, M.; Raghavan, S. DNA double-strand break repair in mammals. J. Radiat. Cancer Res. 2017, 8, 93–97. [Google Scholar]
- Li, Z.; Cloutier, P.; Sanche, L.; Wagner, J.R. Low-energy electron-induced DNA damage: Effect of base sequence in oligonucleotide trimers. J. Am. Chem. Soc. 2010, 132, 5422–5427. [Google Scholar] [CrossRef]
- Keller, A.; Bald, I.; Rotaru, A.; Cauët, E.; Gothelf, K.V.; Besenbacher, F. Probing electron-induced bond cleavage at the single-molecule level using DNA origami templates. ACS Nano 2012, 6, 4392–4399. [Google Scholar] [CrossRef]
- Rackwitz, J.; Bald, I. Low-Energy Electron-Induced Strand Breaks in Telomere-Derived DNA Sequences-Influence of DNA Sequence and Topology. Chem. A Eur. J. 2018, 24, 4680–4688. [Google Scholar] [CrossRef]
- Vogel, S.; Ebel, K.; Schürmann, R.M.; Heck, C.; Meiling, T.; Milosavljevic, A.R.; Giuliani, A.; Bald, I. Vacuum-UV and Low-Energy Electron-Induced DNA Strand Breaks - Influence of the DNA Sequence and Substrate. Chemphyschem 2019, 20, 823–830. [Google Scholar] [CrossRef]
- Solomun, T.; Seitz, H.; Sturm, H. DNA damage by low-energy electron impact: Dependence on guanine content. J. Phys. Chem. B 2009, 113, 11557–11559. [Google Scholar] [CrossRef] [PubMed]
- Huels, M.A.; Boudaïffa, B.; Cloutier, P.; Hunting, D.; Sanche, L. Single, double, and multiple double strand breaks induced in DNA by 3-100 eV electrons. J. Am. Chem. Soc. 2003, 125, 4467–4477. [Google Scholar] [CrossRef] [PubMed]
- Rothemund, P.W.K. Folding DNA to create nanoscale shapes and patterns. Nature 2006, 440, 297–302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banyasz, A.; Balanikas, E.; Martinez-Fernandez, L.; Baldacchino, G.; Douki, T.; Improta, R.; Markovitsi, D. Radicals Generated in Tetramolecular Guanine Quadruplexes by Photoionization: Spectral and Dynamical Features. J. Phys. Chem. B 2019, 123, 4950–4957. [Google Scholar] [CrossRef]
- Banyasz, A.; Ketola, T.; Martínez-Fernández, L.; Improta, R.; Markovitsi, D. Adenine radicals generated in alternating AT duplexes by direct absorption of low-energy UV radiation. Faraday Discuss. 2018, 207, 181–197. [Google Scholar] [CrossRef]
- Simons, J. How do low-energy (0.1–2 eV) electrons cause DNA-strand breaks? Acc. Chem. Res. 2006, 39, 772–779. [Google Scholar] [CrossRef]
- Boulanouar, O.; Fromm, M.; Mavon, C.; Cloutier, P.; Sanche, L. Dissociative electron attachment to DNA-diamine thin films: Impact of the DNA close environment on the OH- and O- decay channels. J. Chem. Phys. 2013, 139, 099901. [Google Scholar] [CrossRef]
- Abdoul-Carime, H.; Cloutier, P.; Sanche, L. Low-Energy (5–40 eV) Electron-Stimulated Desorption of Anions from Physisorbed DNA Bases. Radiat. Res. 2001, 155, 625–633. [Google Scholar] [CrossRef]
- Abdoul-Carime, H.; Dugal, P.-C.; Sanche, L. Damage Induced by 1–30 eV Electrons on Thymine- and Bromouracil-Substituted Oligonucleotides. Radiat. Res. 2000, 153, 23–28. [Google Scholar] [CrossRef]
- Schürmann, R.; Tsering, T.; Tanzer, K.; Denifl, S.; Kumar, S.V.K.; Bald, I. Resonant Formation of Strand Breaks in Sensitized Oligonucleotides Induced by Low-Energy Electrons (0.5–9 eV). Angew. Chem. Int. Ed. Engl. 2017, 56, 10952–10955. [Google Scholar]
- Cauët, E.; Dehareng, D.; Liévin, J. Ab initio study of the ionization of the DNA bases: Ionization potentials and excited states of the cations. J. Phys. Chem. A 2006, 110, 9200–9211. [Google Scholar] [CrossRef] [PubMed]
- Vogel, S. Sequence Dependency of Photon- and Electron Induced DNA Strand Breaks. Chemphyschem 2018, 20, 1–9. [Google Scholar]
- Egli, M.; Saenger, W. Principles of Nucleic Acid Structure; Springer Advanced Texts in Chemistry Ser; Springer: New York, NY, USA, 2013. [Google Scholar]
- Safaee, N.; Noronha, A.M.; Rodionov, D.; Kozlov, G.; Wilds, C.J.; Sheldrick, G.M.; Gehring, K. Structure of the parallel duplex of poly(A) RNA: Evaluation of a 50 year-old prediction. Angew. Chem. Int. Ed. Engl. 2013, 52, 10370–10373. [Google Scholar] [CrossRef] [PubMed]
- Kibbe, W.A. OligoCalc: An online oligonucleotide properties calculator. Nucleic Acids Res. 2007, 35, W43–W46. [Google Scholar] [CrossRef]
- Panajotovic, R.; Martin, F.; Cloutier, P.; Hunting, D.; Sanche, L. Effective cross sections for production of single-strand breaks in plasmid DNA by 0.1 to 4.7 eV electrons. Radiat. Res. 2006, 165, 452–459. [Google Scholar] [CrossRef]
- Boulanouar, O.; Fromm, M.; Bass, A.D.; Cloutier, P.; Sanche, L. Absolute cross section for loss of supercoiled topology induced by 10 eV electrons in highly uniform /DNA/1,3-diaminopropane films deposited on highly ordered pyrolitic graphite. J. Chem. Phys. 2013, 139, 055104. [Google Scholar] [CrossRef] [Green Version]
- Lemelin, V.; Bass, A.D.; Cloutier, P.; Sanche, L. Low energy (1–19 eV) electron scattering from condensed thymidine (dT) II: Comparison of vibrational excitation cross sections with those of tetrahydrofuran and the recalibrated values of thymine. Phys. Chem. Chem. Phys. 2019, 21, 23818–23825. [Google Scholar] [CrossRef]
- Rackwitz, J.; Ranković, M.L.; Milosavljević, A.R.; Bald, I. A novel setup for the determination of absolute cross sections for low-energy electron induced strand breaks in oligonucleotides—The effect of the radiosensitizer 5-fluorouracil*. Eur. Phys. J. D 2017, 71, 32. [Google Scholar] [CrossRef]
- Green, N.M. Avidin. In Advances in Protein Chemistry; Elsevier: Amsterdam, The Netherlands, 1975; Volume 29, pp. 85–133. [Google Scholar]
- Baccarelli, I.; Bald, I.; Gianturco, F.A.; Illenberger, E.; Kopyra, J. Electron-induced damage of DNA and its components: Experiments and theoretical models. Phys. Rep. 2011, 508, 1–44. [Google Scholar] [CrossRef]





| Sequence | ssDNA SB Cross Section σSB [10−15 cm2] | σgeo [10−15 cm2] | |||
|---|---|---|---|---|---|
| 5 eV | 7 eV | 8.4 eV | 10 eV | / | |
| 5′-dA4 | 1.03 ± 0.12 | 4.79 ± 1.04 | 3.27 ± 0.51 | 2.25 ± 0.15 | 11.04 |
| 5′-dA8 | 0.93 ± 0.17 | 5.59 ± 0.66 | 5.09 ± 0.92 | 2.42 ± 0.33 | 22.08 |
| 5′-dA12 | 1.34 ± 0.17 | 6.95 ± 1.12 | 6.32 ± 0.59 | 3.10 ± 0.54 | 33.12 |
| 5′-dA16 | 1.94 ± 0.36 | 7.69 ± 1.02 | 7.62 ± 0.95 | 3.88 ± 0.34 | 44.16 |
| 5′-dA20 | 0.85 ± 0.13 | 5.06 ± 0.28 | 4.72 ± 0.32 | 2.73 ± 0.42 | 55.20 |
| Sequence | SB Cross Section per Nucleotide σN (10−16 cm2) | σN-geo (10−16 cm2) | |||
|---|---|---|---|---|---|
| 5 eV | 7 eV | 8.4 eV | 10 eV | / | |
| 5′-dA4 | 2.58 | 11.98 | 8.18 | 5.63 | 27.60 |
| 5′-dA8 | 1.16 | 6.99 | 6.36 | 3.03 | 27.60 |
| 5′-dA12 | 1.12 | 5.79 | 5.27 | 2.58 | 27.60 |
| 5′-dA16 | 1.21 | 4.81 | 4.76 | 2.43 | 27.60 |
| 5′-dA20 | 0.43 | 2.53 | 2.36 | 1.37 | 27.60 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ebel, K.; Bald, I. Length and Energy Dependence of Low-Energy Electron-Induced Strand Breaks in Poly(A) DNA. Int. J. Mol. Sci. 2020, 21, 111. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21010111
Ebel K, Bald I. Length and Energy Dependence of Low-Energy Electron-Induced Strand Breaks in Poly(A) DNA. International Journal of Molecular Sciences. 2020; 21(1):111. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21010111
Chicago/Turabian StyleEbel, Kenny, and Ilko Bald. 2020. "Length and Energy Dependence of Low-Energy Electron-Induced Strand Breaks in Poly(A) DNA" International Journal of Molecular Sciences 21, no. 1: 111. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21010111