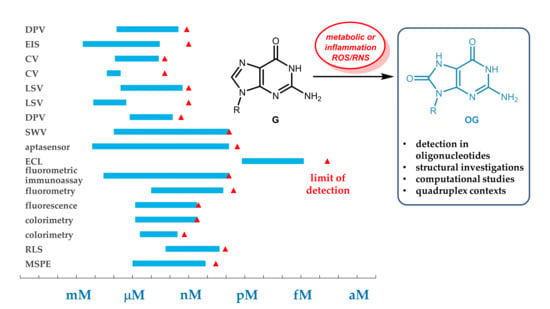
Analytical and Structural Studies for the Investigation of Oxidative Stress in Guanine Oligonucleotides
Abstract
:1. Introduction
2. Scope and Limitations
3. Analytical Detection and Quantitation Methods
3.1. The Concentration of ROS, Reactive Nitrogen Species (RNS) and the Characteristic Processes Leading to Their Formation
3.2. Electrochemical Methods
3.2.1. Development of New Electrodes
Electrodes Coated with Reduced Graphene Oxide Nanocomposites
GCE Coated with Multi-Walled Carbon Nanotubes (MWCNTs)
GCE Coated with Porous Single-Walled Carbon Nanotube (PSWCNT)
Voltammetric Sensor for Oxidized DNA Using Ultrathin Films of Osmium and Ruthenium Metallopolymers
Copper-Based Metal Organic Framework Nanoparticles Anchored to Graphite Nanosheets
3.2.2. Gold Electrodes with o8dGuo-Specific Aptamers
3.2.3. Electrochemiluminescence in a Multiple-Mechanism-Driven Biosensor
3.3. Optical Methods
3.3.1. Fluorometric Assays
3.3.2. Visual Analysis of Samples without the Use of Expensive Instruments
3.3.3. Resonance Light Scattering Aptasensor Based on Magnetic Nanoparticles
3.3.4. UV Radiation
3.4. Hyphenated Techniques
3.4.1. Gas Chromatography–Mass Spectrometry (GC–MS) Analyses
3.4.2. G Oxidation Study by EC-LC-MS Chromatography Technique
3.4.3. Aptamer-Based Online Magnetic Solid Phase Extraction for HPLC–MS Analysis
3.5. Summary and Comparison of the Efficacy of Analytical Methods
4. Structural Studies and Assays for Investigation of Structure–Function Relationship
4.1. Nanopore Detection
4.2. NMR and CD Spectroscopy and Tm Studies to Investigate the Secondary and 3D Structure of GQs
4.2.1. NMR Spectroscopy
4.2.2. CD Spectroscopy
4.2.3. Melting Temperature (Tm) Analysis and Its Combination with 8-Bromoguanine Scan
4.3. Functional in vitro and in vivo Assays to Understand the Role of Oxidative Damage of GQs in Gene Regulation
4.3.1. The Response of GQs to Oxidative Damage
4.3.2. OG–Xanthine Tetrads in Quadruplex Structures
4.4. Computational Simulations of Quadruplexes: Effects of OG Incorporation or Apurinic/Apyrimidinic Sites
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| 2Ih | 5-carboxamido-5-formamido-2-iminohydantoin |
| α-HL | α-hemolysin |
| AuNP | gold nanoparticles |
| bpy | 2:2’-bipyridine |
| C | cytosine |
| c7G | 7-deazaguanine |
| CD | circular dichroism |
| CNNS | carbon nitride nanosheets |
| CQD | carbon quantum dot |
| CV | cyclic voltammetry |
| DA | dopamine |
| dG | 2’-deoxyguanosine in ON sequences (the prefix d in front of parenthesis relates to the whole sequence) |
| DMAc | N,N-dimethylacetamide |
| DPV | differential pulse voltammetry |
| (d)Ribf | (2-deoxy-)b-d-ribofuranosyl |
| dsDNA | double-stranded DNA |
| DW | DNAwalker |
| EC-LC-MS | electrochemistry-liquid chromatography-mass spectrometry |
| ECD | electrochemical detection |
| ECL | electrochemiluminescence |
| EIS | electrochemical impedance spectroscopy |
| Fapy-G | 2,6-diamino-4-hydroxy-5-formamidopyrimidine |
| G | guanine (in ON sequences guanosine) |
| G4 | guanine tetrad |
| GCE | glassy carbon electrode |
| Gh | 5-guanidinohydantoin |
| Ghox | 5-guanidinodehydrohydantoin |
| GN | graphite nanosheets |
| GO | graphene oxide |
| GQ | guanine quadruplex |
| HCR | hybridization chain reaction |
| hm5Ura | 5-hydroxymethyluracil |
| HPLC | high-performance liquid chromatography |
| hTel | human telomere |
| Iz | 2,5-diaminoimidazolone |
| LOD | limit of detection |
| LSV | linear sweep voltammetry |
| m5C | 5-methylcytosine |
| MB | methylene blue |
| MCH | 6-mercapto-1-hexanol |
| MNP | magnetic nanoparticles |
| MWCNT | multi-walled carbon nanotubes |
| NEase | nicking endonuclease |
| NEIL3 | Nei-like or endonuclease VIII-like 3 enzyme |
| NHE | normal hydrogen electrode |
| NMR | nuclear magnetic resonance |
| NOE | nuclear Overhauser effect |
| nt | nucleotide |
| o8dGuo | 8-oxo-7,8-dihydro-2′-deoxyguanosine |
| OG | 8-oxo-7,8-dihydroguanine |
| OGG1 | OG-glycosylase 1 |
| ON | oligonucleotide |
| PSWCNT | porous single-walled carbon nanotube |
| PT | poly(3-alkoxy-4-methylthiophene) |
| PVP | poly(vinylpyridines) |
| QCM | quartz crystal microbalance |
| rGO | reduced graphene oxide |
| RAD17 | the gene encoding cell cycle checkpoint protein RAD17 |
| RLS | resonance light scattering |
| RNS | reactive nitrogen species |
| ROS | reactive oxygen species |
| SEM | scanning electron microscopy |
| S/N | signal-to-noise ratio |
| SOD | superoxide dismutase |
| Sp | spiroiminodihydantoin |
| SPCE | screen-printed carbon electrode |
| ssDNA | single-stranded DNA |
| SWCNT | single-walled carbon nanotube |
| SWV | square wave voltammetry |
| TD-DFT | time-dependent density-functional theory |
| TEM | transmission electron microscopy |
| TGA | thermogravimetric analysis |
| UA | uric acid |
| XRD | X-ray diffractometry |
| Z | 2,2,4-triamino-2H-oxazol-5-one |
References
- Rigo, R.; Palumbo, M.; Sissi, C. G-quadruplexes in human promoters: A challenge for therapeutic applications. Biochim. Biophys. Acta 2017, 1861, 1399–1413. [Google Scholar] [CrossRef] [PubMed]
- Szalai, V.A.; Singer, M.J.; Thorp, H.H. Site-specific probing of oxidative reactivity and telomerase function using 7,8-dihydro-8-oxoguanine in telomeric DNA. J. Am. Chem. Soc. 2002, 124, 1625–1631. [Google Scholar] [CrossRef] [PubMed]
- Omaga, C.A.; Fleming, A.M.; Burrows, C.J. The fifth domain in the G-quadruplex-forming sequence of the human NEIL3 promoter locks DNA folding in response to oxidative damage. Biochemistry 2018, 57, 2958–2970. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Ding, P.; Xie, C.; Ye, C.; Ye, M.; Pan, C.; Cao, X.; Zhang, S.; Zheng, S. Potential application of the oxidative nucleic acid damage biomarkers in detection of diseases. Oncotarget 2017, 8, 75767–75777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fleming, A.M.; Burrows, C.J. Interplay of guanine oxidation and G-quadruplex folding in gene promoters. J. Am. Chem. Soc. 2020, 142, 1115–1136. [Google Scholar] [CrossRef]
- Fleming, A.M.; Zhu, J.; Ding, Y.; Burrows, C.J. 8-Oxo-7,8-dihydroguanine in the context of a gene promoter G-quadruplex is an on–off switch for transcription. ACS Chem. Biol. 2017, 12, 2417–2426. [Google Scholar] [CrossRef]
- Giorgio, M.; Dellino, G.I.; Gambino, V.; Roda, N.; Pelicci, P.G. On the epigenetic role of guanosine oxidation. Redox Biol. 2020, 29, 101398:1–101398:14. [Google Scholar] [CrossRef]
- Markkanen, E. Not breathing is not an option: How to deal with oxidative DNA damage. DNA Repair 2017, 59, 82–105. [Google Scholar] [CrossRef]
- Fleming, A.M.; Ding, Y.; Burrows, C.J. Oxidative DNA damage is epigenetic by regulating gene transcription via base excision repair. Proc. Natl. Acad. Sci. USA 2017, 114, 2604–2609. [Google Scholar] [CrossRef]
- Fleming, A.M.; Burrows, C.J. 8-Oxo-7,8-dihydroguanine, friend and foe: Epigenetic-like regulator versus initiator of mutagenesis. DNA Repair 2017, 56, 75–83. [Google Scholar] [CrossRef]
- Sagi, J. In what ways do synthetic nucleotides and natural base lesions alter the structural stability of G-quadruplex nucleic acids? J. Nucl. Acids 2017, 1641845:1–1641845:45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ba, X.; Boldogh, I. 8-Oxoguanine DNA glycosylase 1: Beyond repair of the oxidatively modified base lesions. Redox Biol. 2018, 14, 669–678. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, W.; Lingner, J. Impact of oxidative stress on telomere biology. Differentiation 2018, 99, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Spiegel, J.; Adhikari, S.; Balasubramanian, S. The structure and function of DNA G-quadruplexes. Trends Chem. 2020, 2, 123–136. [Google Scholar] [CrossRef] [Green Version]
- Tavakoli, P.; Leifert, W.; Fenech, M.; François, M. Guanine-quadruplexes and possible role in nutritional epigenetics and aging. In Handbook of Nutrition, Diet, and Epigenetics; Patel, V.B., Preedy Victor, R., Eds.; Springer: Cham, Switzerland, 2019; pp. 293–309. ISBN 978-3-319-55529-4. [Google Scholar]
- Bokhari, B.; Sharma, S. Stress marks on the genome: Use or lose? Int. J. Mol. Sci. 2019, 20, 364. [Google Scholar] [CrossRef] [Green Version]
- François, M.; Leifert, W.; Tellam, R.; Fenech, M. G-quadruplexes: A possible epigenetic target for nutrition. Mutat. Res. Rev. Mutat. Res. 2015, 764, 101–107. [Google Scholar] [CrossRef]
- Fleming, A.M.; Burrows, C.J.; AF Fleming, A.M.; Burrows, C.J. G-quadruplex folds of the human telomere sequence alter the site reactivity and reaction pathway of guanine oxidation compared to duplex DNA. Chem. Res. Toxicol. 2013, 26, 593–607. [Google Scholar] [CrossRef] [Green Version]
- Cadet, J.; Douki, T.; Gasparutto, D.; Ravanat, J.-L.; Wagner, J.R. Oxidatively generated nucleobase modifications in isolated and cellular DNA. In Encyclopedia of Radicals in Chemistry, Biology and Materials; Chatgilialoglu, C., Studer, A., Eds.; John Wiley and Sons: Chichester, NJ, USA, 2012; pp. 985–1010. ISBN 9781119953678. [Google Scholar]
- Geacintov, N.E.; Broyde, S. The Chemical Biology of DNA Damage; Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2010; ISBN 3527322957. [Google Scholar]
- Cadet, J.; Wagner, J.R. DNA base damage by reactive oxygen species, oxidizing agents, and UV radiation. CSH Perspect. Biol. 2013, 5, a012559:1–a012559:18. [Google Scholar] [CrossRef]
- Pham, A.N.; Xing, G.; Miller, C.J.; Waite, T.D. Fenton-like copper redox chemistry revisited: Hydrogen peroxide and superoxide mediation of copper-catalyzed oxidant production. J. Catal. 2013, 301, 54–64. [Google Scholar] [CrossRef]
- Illés, E.; Mizrahi, A.; Marks, V.; Meyerstein, D. Carbonate-radical-anions, and not hydroxyl radicals, are the products of the Fenton reaction in neutral solutions containing bicarbonate. Free Radical Biol. Med. 2019, 131, 1–6. [Google Scholar] [CrossRef]
- Heller, A. Spiers Memorial Lecture. On the hypothesis of cathodic protection of genes. Faraday Discuss. 2000, 116, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Sistare, M.F.; Codden, S.J.; Heimlich, G.; Thorp, H.H. Effects of base stacking on guanine electron transfer: Rate constants for G and GG sequences of oligonucleotides from catalytic electrochemistry. J. Am. Chem. Soc. 2000, 122, 4742–4749. [Google Scholar] [CrossRef]
- Hao, J.; Wu, K.; Wan, C.; Tang, Y. Reduced graphene oxide-ZnO nanocomposite based electrochemical sensor for sensitive and selective monitoring of 8-hydroxy-2’-deoxyguanosine. Talanta 2018, 185, 550–556. [Google Scholar] [CrossRef]
- Manavalan, S.; Rajaji, U.; Chen, S.-M.; Steplin Paul Selvin, S.; Govindasamy, M.; Chen, T.-W.; Ajmal Ali, M.; Al-Hemaid, F.M.A.; Elshikh, M.S. Determination of 8-hydroxy-2′-deoxyguanosine oxidative stress biomarker using dysprosium oxide nanoparticles@reduced graphene oxide. Inorg. Chem. Front. 2018, 5, 2885–2892. [Google Scholar] [CrossRef]
- Guo, Z.; Liu, X.; Liu, Y.; Wu, G.; Lu, X. Constructing a novel 8-hydroxy-2′-deoxyguanosine electrochemical sensor and application in evaluating the oxidative damages of DNA and guanine. Biosens. Bioelectron. 2016, 86, 671–676. [Google Scholar] [CrossRef]
- Shang, T.; Wang, P.; Liu, X.; Jiang, X.; Hu, Z.; Lu, X. Facile synthesis of porous single-walled carbon nanotube for sensitive detection of 8-hydroxy-2′-deoxyguanosine. J. Electroanal. Chem. 2018, 808, 28–34. [Google Scholar] [CrossRef]
- Mugweru, A.; Wang, B.Q.; Rusling, J. Voltammetric sensor for oxidized DNA using ultrathin films of osmium and ruthenium metallopolymers. Anal. Chem. 2004, 76, 5557–5563. [Google Scholar] [CrossRef] [PubMed]
- Dennany, L.; Forster, R.J.; White, B.; Smyth, M.; Rusling, J.F. Direct electrochemiluminescence detection of oxidized DNA in ultrathin films containing [Os(bpy)2(PVP)10]2+. J. Am. Chem. Soc. 2004, 126, 8835–8841. [Google Scholar] [CrossRef]
- Cao, G.; Wu, C.; Tang, Y.; Wan, C. Ultrasmall HKUST-1 nanoparticles decorated graphite nanosheets for highly sensitive electrochemical sensing of DNA damage biomarker 8-hydroxy-2’-deoxyguanosine. Anal. Chim. Acta 2019, 1058, 80–88. [Google Scholar] [CrossRef]
- Zheng, J.; Gao, T.; Shi, H.; Huang, Y.; Xiang, Y.; Li, G. Electrochemical analysis of 8-hydroxy-2’-deoxyguanosine with enhanced sensitivity based on exonuclease-mediated functional nucleic acid. Talanta 2019, 199, 324–328. [Google Scholar] [CrossRef]
- Jia, L.-P.; Wang, L.-J.; Ma, R.N.; Shang, L.; Zhang, W.; Xue, Q.-W.; Wang, H.-S. An electrochemical aptasensor for the highly sensitive detection of 8-hydroxy-2′-deoxyguanosine based on the hybridization chain reaction. Talanta 2018, 179, 414–419. [Google Scholar] [CrossRef]
- Lv, Y.; Chen, S.; Shen, Y.; Ji, J.; Zhou, Q.; Liu, S.; Zhang, Y. Competitive multiple-mechanism-driven electrochemiluminescent detection of 8-hydroxy-2’-deoxyguanosine. J. Am. Chem. Soc. 2018, 140, 2801–2804. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Chen, T.; Zhang, Y.; Ye, W.W.; AF Hu, W.; Chen, T.; Zhang, Y.; Ye, W. A carbon dot and gold nanoparticle-based fluorometric immunoassay for 8-hydroxy-2’-deoxyguanosine in oxidatively damaged DNA. Microchim. Acta 2019, 186, 303:1–303:9. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Wei, M.; Yin, L.; Pu, Y.; Liu, S. Improving the fluorometric determination of the cancer biomarker 8-hydroxy-2′-deoxyguanosine by using a 3D DNA nanomachine. Microchim. Acta 2018, 185, 494:1–494:7. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.-C.; Wang, Y.-S.; Rang, W.-Q.; Xue, J.-H.; Zhou, B.; Liu, L.; Qian, Q.-M.; Wang, Y.-S.; Yin, J.-C. Colorimetric determination of 8-hydroxy-2’-deoxyguanosine using label-free aptamer and unmodified gold nanoparticles. Microchim. Acta 2014, 181, 903–910. [Google Scholar] [CrossRef]
- Ammanath, G.; Yildiz, U.H.; Palaniappan, A.; Liedberg, B. Luminescent device for the detection of oxidative stress biomarkers in artificial urine. ACS Appl. Mater. Interfaces 2018, 10, 7730–7736. [Google Scholar] [CrossRef]
- Tao, L.X.; Yue, Q.L.; Hou, Y.N.; Wang, Y.P.; Chen, C.Y.; Li, C.Z.; AF Tao, L.; Yue, Q.; Hou, Y.; Wang, Y.; et al. Resonance light scattering aptasensor for urinary 8-hydroxy-2’-deoxyguanosine based on magnetic nanoparticles: A preliminary study of oxidative stress association with air pollution. Microchim. Acta 2018, 185, 419:1–419:7. [Google Scholar] [CrossRef]
- Banyasz, A.; Martinez-Fernandez, L.; Balty, C.; Perron, M.; Douki, T.; Improta, R.; Markovitsi, D. Absorption of low-energy UV radiation by human telomere G-quadruplexes generates long-lived guanine radical cations. J. Am. Chem. Soc. 2017, 139, 10561–10568. [Google Scholar] [CrossRef]
- Gomez-Mendoza, M.; Banyasz, A.; Douki, T.; Markovitsi, D.; Ravanat, J.-L. Direct oxidative damage of naked DNA generated upon absorption of UV radiation by nucleobases. J. Phys. Chem. Lett. 2016, 7, 3945–3948. [Google Scholar] [CrossRef]
- Rozalski, R.; Gackowski, D.; Siomek-Gorecka, A.; Starczak, M.; Modrzejewska, M.; Banaszkiewicz, Z.; Olinski, R. Urinary 5-hydroxymethyluracil and 8-oxo-7,8-dihydroguanine as potential biomarkers in patients with colorectal cancer. Biomarkers 2015, 20, 287–291. [Google Scholar] [CrossRef]
- Oberacher, H.; Erb, R.; Plattner, S.; Chervet, J.P.; AF Oberacher, H.; Erb, R.; Plattner, S.; Chervet, J.-P. Mechanistic aspects of nucleic-acid oxidation studied with electrochemistry-mass spectrometry. Trends Anal. Chem. 2015, 70, 100–111. [Google Scholar] [CrossRef]
- Gan, H.; Xu, H. A novel aptamer-based online magnetic solid phase extraction method for the selective determination of 8-hydroxy-2′-deoxyguanosine in human urine. Anal. Chim. Acta 2018, 1008, 48–56. [Google Scholar] [CrossRef]
- An, N.; Fleming, A.M.; White, H.S.; Burrows, C.J. Nanopore detection of 8-oxoguanine in the human telomere repeat sequence. ACS Nano 2015, 9, 4296–4307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, Y.; Fleming, A.M.; White, H.S.; Burrows, C.J. Differentiation of G:C vs A:T and G:C vs G:mC base pairs in the latch zone of α-hemolysin. ACS Nano 2015, 9, 11325–11332. [Google Scholar] [CrossRef] [Green Version]
- Jin, Q.; Fleming, A.M.; Johnson, R.P.; Ding, Y.; Burrows, C.J.; White, H.S. Base-excision repair activity of uracil-DNA glycosylase monitored using the latch zone of α-hemolysin. J. Am. Chem. Soc. 2013, 135, 19347–19353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, R.P.; Fleming, A.M.; Burrows, C.J.; White, H.S. Effect of an electrolyte cation on detecting DNA damage with the latch constriction of α-hemolysin. J. Phys. Chem. Lett. 2014, 5, 3781–3786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, R.; Fleming, A.; Jin, Q.; Burrows, C.; White, H. Temperature and electrolyte optimization of the α-hemolysin latch sensing zone for detection of base modification in double-stranded DNA. Biophys. J. 2014, 107, 924–931. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, T.; Yang, J.; Pan, V.; Zhao, N.; Lu, Z.; Ke, Y.; Zhang, C. DNA nanotechnology assisted nanopore-based analysis. Nucleic Acids Res. 2020, 48, 2791–2806. [Google Scholar] [CrossRef] [Green Version]
- Tang, F.; Liu, S.; Li, Q.-Y.; Yuan, J.; Li, L.; Wang, Y.; Yuan, B.-F.; Feng, Y.Q. Location analysis of 8-oxo-7,8-dihydroguanine in DNA by polymerase-mediated differential coding. Chem. Sci. 2019, 10, 4272–4281. [Google Scholar] [CrossRef] [Green Version]
- Zhu, J.; Fleming, A.M.; Burrows, C.J. The RAD17 promoter sequence contains a potential tail-dependent G-quadruplex that downregulates gene expression upon oxidative modification. ACS Chem. Biol. 2018, 13, 2577–2584. [Google Scholar] [CrossRef]
- Lim, K.W.; Lacroix, L.; Yue, D.J.E.; Lim, J.K.C.; Lim, J.M.W.; Phan, A.T. Coexistence of two distinct G-quadruplex conformations in the hTERT promoter. J. Am. Chem. Soc. 2010, 132, 12331–12342. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Dexheimer, T.S.; Chen, D.; Carver, M.; Ambrus, A.; Jones, R.A.; Yang, D. An intramolecular G-quadruplex structure with mixed parallel/antiparallel G-strands formed in the human Bcl-2 promoter region in solution. J. Am. Chem. Soc. 2006, 128, 1096–1098. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mangerich, A.; Knutson, C.G.; Parry, N.M.; Muthupalani, S.; Ye, W.; Prestwich, E.; Cui, L.; McFaline, J.L.; Mobley, M.; Ge, Z.; et al. Infection-induced colitis in mice causes dynamic and tissue-specific changes in stress response and DNA damage leading to colon cancer. Proc. Natl. Acad. Sci. USA 2012, 109, E1820–E1829. [Google Scholar] [CrossRef] [Green Version]
- Cogoi, S.; Ferino, A.; Miglietta, G.; Pedersen, E.B.; Xodo, L.E. The regulatory G4 motif of the Kirsten ras (KRAS) gene is sensitive to guanine oxidation: Implications on transcription. Nucleic Acids Res. 2018, 46, 661–676. [Google Scholar] [CrossRef] [PubMed]
- Müller, G.A.; Engeland, K. The central role of CDE/CHR promoter elements in the regulation of cell cycle-dependent gene transcription. FEBS J. 2010, 277, 877–893. [Google Scholar] [CrossRef] [PubMed]
- Neurauter, C.G.; Luna, L.; Bjørås, M. Release from quiescence stimulates the expression of human NEIL3 under the control of the Ras dependent ERK–MAP kinase pathway. DNA Repair 2012, 11, 401–409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olsen, C.M.; Gmeiner, W.H.; Marky, L.A. Unfolding of G-Quadruplexes: energetic, and ion and water contributions of G-quartet stacking. J. Phys. Chem. B 2006, 110, 6962–6969. [Google Scholar] [CrossRef]
- Benz, A.; Hartig, J.S. Redesigned tetrads with altered hydrogen bonding patterns enable programming of quadruplex topologies. Chem. Commun. 2008, 4010–4012. [Google Scholar] [CrossRef] [Green Version]
- Paragi, G.; Fonseca Guerra, C. Cooperativity in the self-assembly of the guanine nucleobase into quartet and ribbon structures on surfaces. Chem. Eur. J. 2017, 23, 3042–3050. [Google Scholar] [CrossRef]
- Cheong, V.V.; Heddi, B.; Lech, C.J.; Phan, A.T. Xanthine and 8-oxoguanine in G-quadruplexes: Formation of a G· G· X· O tetrad. Nucleic Acids Res. 2015, 43, 10506–10514. [Google Scholar] [CrossRef] [Green Version]
- Cheong, V.V.; Lech, C.J.; Heddi, B.; Phan, A.T. Inverting the G-tetrad polarity of a G-quadruplex by using xanthine and 8-oxoguanine. Angew. Chem. Int. Ed. 2016, 55, 160–163. [Google Scholar] [CrossRef] [PubMed]
- Fujii, T.; Podbevsek, P.; Plavec, J.; Sugimoto, N. Effects of metal ions and cosolutes on G-quadruplex topology. J. Inorg. Biochem. 2017, 166, 190–198. [Google Scholar] [CrossRef] [PubMed]
- Dumont, E.; Grüber, R.; Bignon, E.; Morell, C.; Moreau, Y.; Monari, A.; Ravanat, J.-L. Probing the reactivity of singlet oxygen with purines. Nucleic Acids Res. 2016, 44, 56–62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bielskute, S.; Plavec, J.; Podbevsek, P. Impact of oxidative lesions on the human telomeric G-quadruplex. J. Am. Chem. Soc. 2019, 141, 2594–2603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hognon, C.; Gebus, A.; Barone, G.; Monari, A. Human DNA Telomeres in presence of oxidative lesions: The crucial role of electrostatic interactions on the stability of guanine quadruplexes. Antioxidants 2019, 8, 337. [Google Scholar] [CrossRef] [Green Version]




















| Method(s) Used 1 | Compound Detected | Specificity and Other Characteristic(s) | Reference |
|---|---|---|---|
| CV, DPV | o8dGuo | uricase treatment eliminated UA interference | Hao et al., 2018 [26] |
| EIS | o8dGuo | the UA peak potential was different (+0.18 V) and of lower intensity than that of o8dGuo (+0.37 V) | Manavalan et al., 2018 [27] |
| EIS, CV | o8dGuo | well-separated oxidation peaks for o8dGuo, UA and DA | Guo et al., 2016 [28] |
| LSV | o8dGuo | temperature-dependent, kinetic and thermodynamic parameters have been obtained as well | Shang et al., 2018 [29] |
| SWV | DNA | a single oxidized nucleobase in 6000 can be detected in thin metallopolymer films | Mugweru et al., 2004 [30] |
| ECL | DNA | thin metallopolymer films can detect OG lesion in DNA | Dennany et al., 2004 [31] |
| DPV | o8dGuo | uricase treatment eliminated UA interference | Cao et al., 2019 [32] |
| EIS, CV, SWV | o8dGuo | o8dGuo–aptamer–DNAzyme complex | Zheng et al., 2019 [33] |
| aptasensor, EIS, DPV | DNA | HCR-enhanced detection of oxidative lesion using GQ formation | Jia et al., 2018 [34] |
| ECL | o8dGuo | multiple-mechanism-driven sensor based on GQ/hemin aptamer; extreme sensitivity | Lv et al., 2018 [35] |
| fluorometric immunoassay | DNA | it requires o8dGuo isolated from damaged DNA | Hu et al., 2019 [36] |
| fluorometry | o8dGuo | AuNP-attached 3D DNA nanomachine | Wei et al., 2018 [37] |
| colorimetry, CD | o8dGuo | naked eye detection of o8dGuo–GQ-aptamer | Wang et al., 2014 [38] |
| luminescence, colorimetry | o8dGuo | naked eye detection of o8dGuo–GQ-aptamer | Ammanath et al., 2018 [39] |
| RLS | o8dGuo | o8dGuo–GQ-aptamer, MNPs | Tao et al., 2018 [40] |
| nanosecond time-resolved UV spectroscopy | DNA GQ | transient guanine radicals appear as precursors to oxidative damage, generated by absorption of UV radiation | Banyasz et al., 2017 [41] |
| GC–MS | OG | moderate diagnostic performance | Rozalski et al., 2015 [43] |
| EC–LC–MS | G, o8(d)Guo, short DNA | inter-strand and intra-strand cross-links have been observed beyond oxidation upon EC interaction | Oberacher et al., 2015 [44] |
| MSPE, HPLC–MS | o8dGuo | aptamer-based sample concentration using magnetic forces; enhanced sensitivity | Gan et al., 2018 [45] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ferenc, G.; Váradi, Z.; Kupihár, Z.; Paragi, G.; Kovács, L. Analytical and Structural Studies for the Investigation of Oxidative Stress in Guanine Oligonucleotides. Int. J. Mol. Sci. 2020, 21, 4981. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21144981
Ferenc G, Váradi Z, Kupihár Z, Paragi G, Kovács L. Analytical and Structural Studies for the Investigation of Oxidative Stress in Guanine Oligonucleotides. International Journal of Molecular Sciences. 2020; 21(14):4981. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21144981
Chicago/Turabian StyleFerenc, Györgyi, Zoltán Váradi, Zoltán Kupihár, Gábor Paragi, and Lajos Kovács. 2020. "Analytical and Structural Studies for the Investigation of Oxidative Stress in Guanine Oligonucleotides" International Journal of Molecular Sciences 21, no. 14: 4981. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21144981