Review of PIP2 in Cellular Signaling, Functions and Diseases
Abstract
:1. Introduction
2. PIP2 in Actin Dynamics
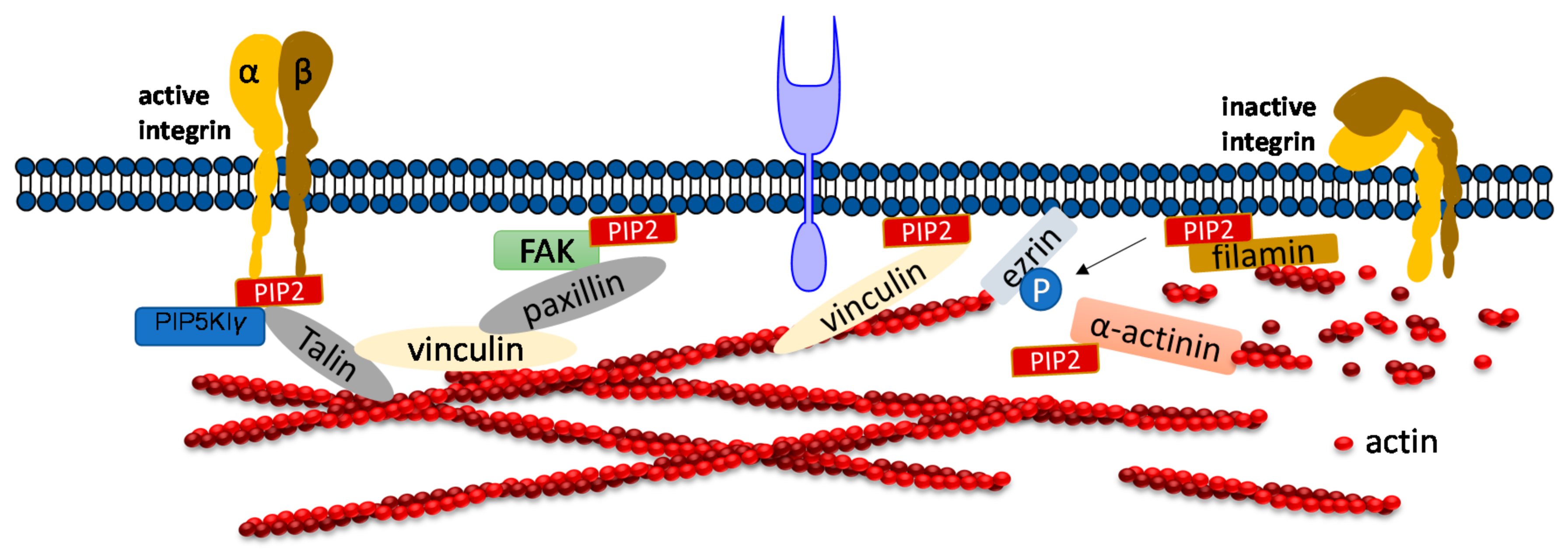
3. PIP2 in Adhesion Dynamics
4. PIP2 in Membrane Dynamics and Organization
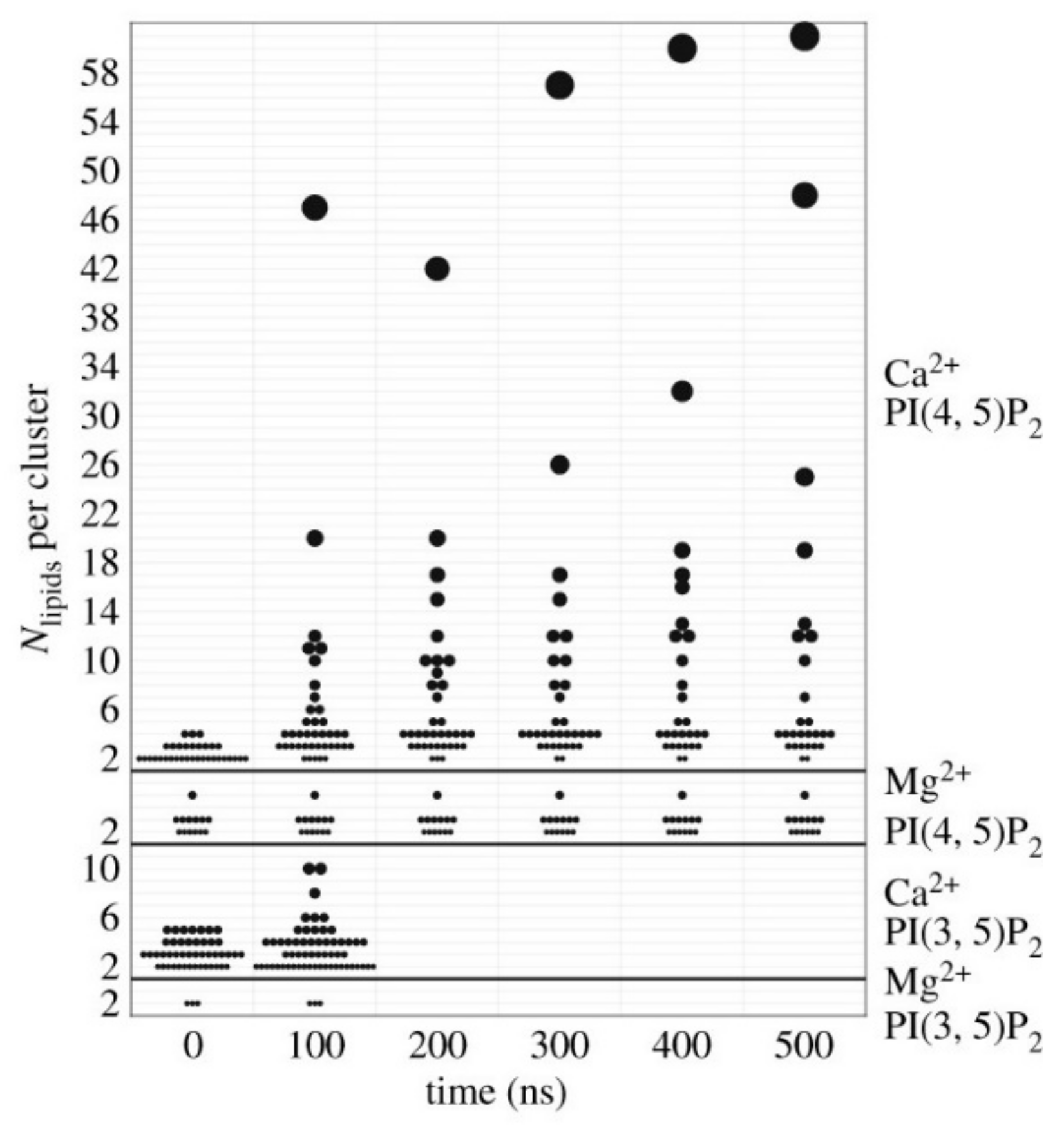
4.1. Charge Dependence and Electrostatic Interaction
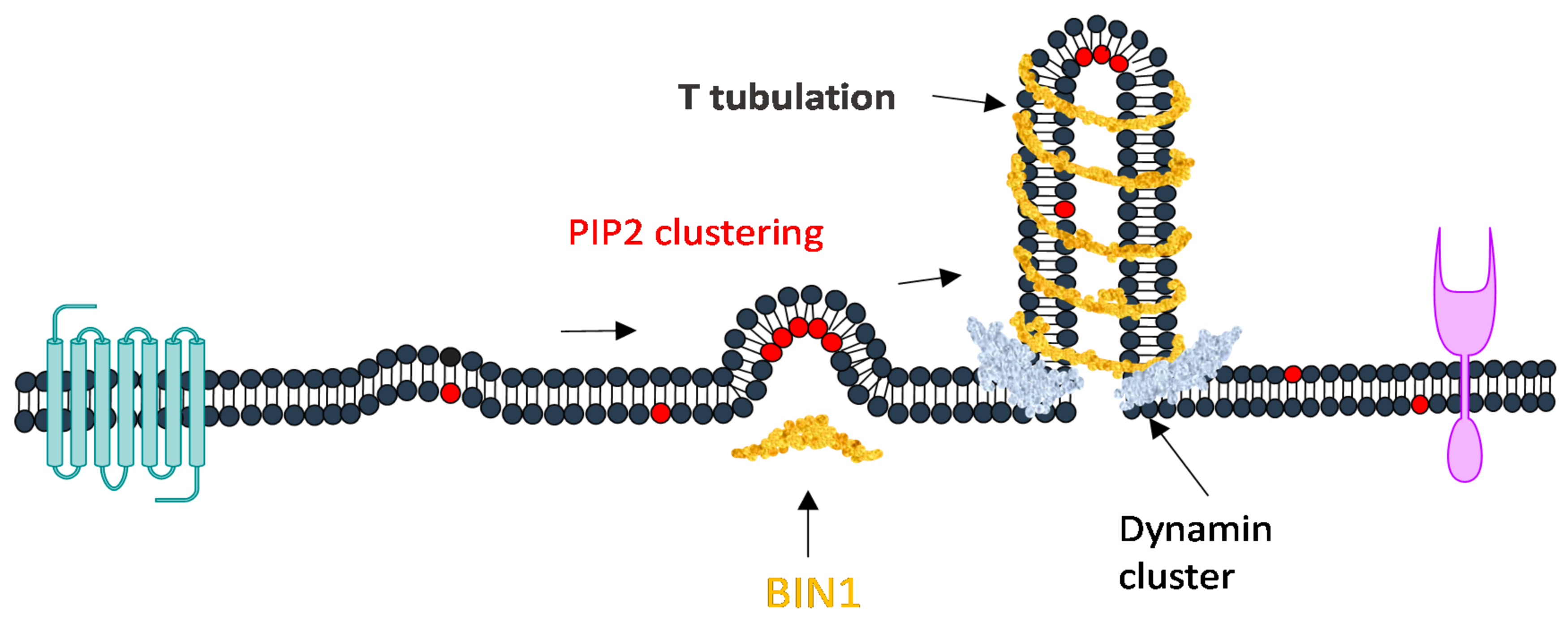
4.2. PIP2 Regulation in Membrane Curvature Sensing and Transport
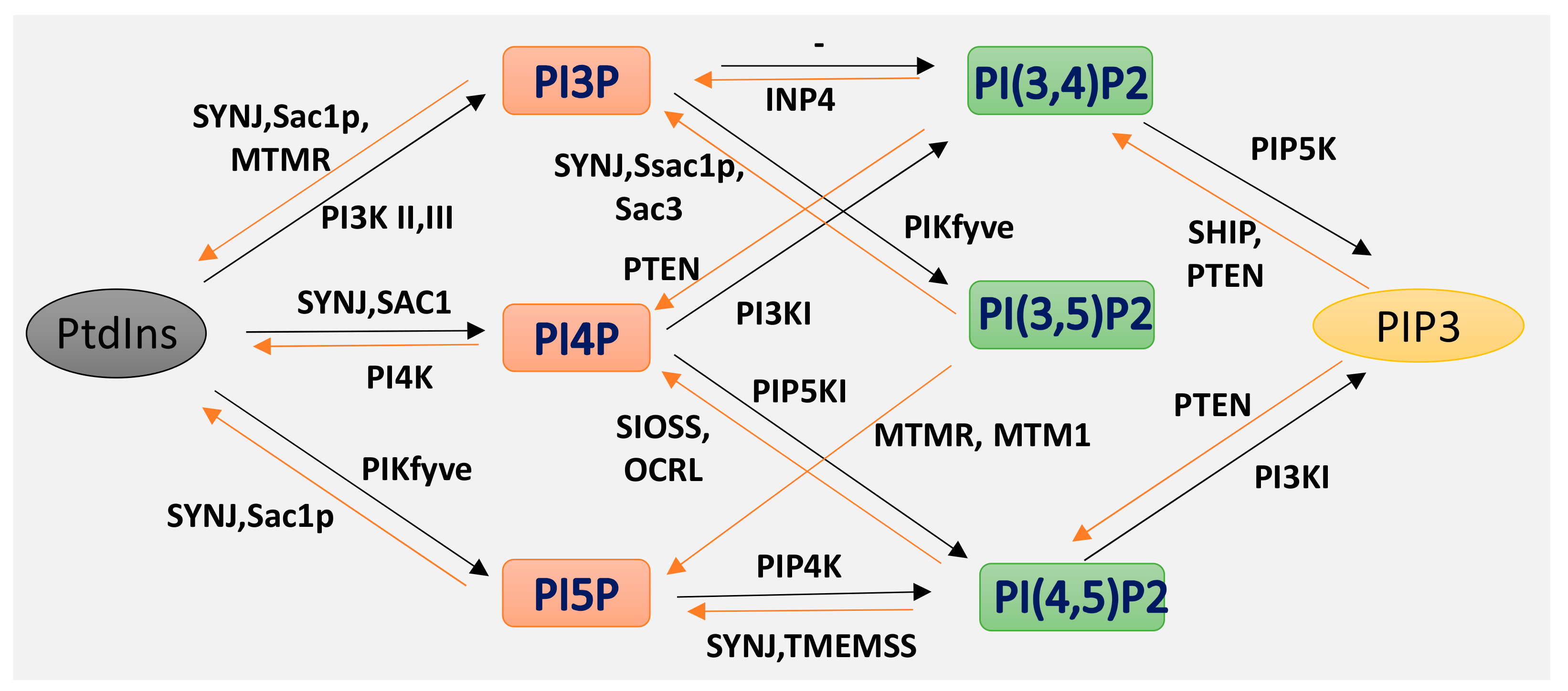
5. Intracellular Trafficking
6. PIP2 in Signaling and Diseases
7. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Downes, C.P.; Gray, A.; Lucocq, J.M. Probing phosphoinositide functions in signaling and membrane trafficking. Trends Cell Biol. 2005, 15, 259–268. [Google Scholar]
- Picas, L.; Viaud, J.; Schauer, K.; Vanni, S.; Hnia, K.; Fraisier, V.; Roux, A.; Bassereau, P.; Gaits-Iacovoni, F.; Payrastre, B.; et al. BIN1/M-Amphiphysin2 induces clustering of phosphoinositides to recruit its downstream partner dynamin. Nat. Commun. 2014, 5, 1–12. [Google Scholar] [CrossRef] [Green Version]
- McLean, M.A.; Stephen, A.G.; Sligar, S.G. PIP2 Influences the Conformational Dynamics of Membrane-Bound KRAS4b. Biochemistry 2019, 58, 3537–3545. [Google Scholar] [CrossRef] [PubMed]
- Romarowski, A.; Battistone, M.A.; La Spina, F.A.; del Puga Molina, L.C.; Luque, G.M.; Vitale, A.M.; Cuasnicu, P.S.; Visconti, P.E.; Krapf, D.; Buffone, M.G. PKA-dependent phosphorylation of LIMK1 and Cofilin is essential for mouse sperm acrosomal exocytosis. Dev. Biol. 2015, 405, 237–249. [Google Scholar] [CrossRef] [Green Version]
- Liepiņa, I.; Czaplewski, C.; Janmey, P.; Liwo, A. Molecular dynamics study of a gelsolin-derived peptide binding to a lipid bilayer containing phosphatidylinositol 4,5-bisphosphate. Biopolymers 2003, 71, 49–70. [Google Scholar] [CrossRef] [PubMed]
- Amano, T.; Tanabe, K.; Eto, T.; Narumiya, S.; Mizuno, K. LIM-kinase 2 induces formation of stress fibres, focal adhesions and membrane blebs, dependent on its activation by Rho-associated kinase-catalysed phosphorylation at threonine-505. Biochem. J. 2001, 354, 149–159. [Google Scholar] [CrossRef] [PubMed]
- Walter, L.M.; Franz, P.; Lindner, R.; Tsiavaliaris, G.; Hensel, N.; Claus, P. Profilin2a-phosphorylation as a regulatory mechanism for actin dynamics. FASEB J. 2020, 34, 2147–2160. [Google Scholar] [CrossRef] [Green Version]
- Saltel, F.; Mortier, E.; Hytönen, V.P.; Jacquier, M.C.; Zimmermann, P.; Vogel, V.; Liu, W.; Wehrle-Haller, B. New PI(4,5)P2- and membrane proximal integrin-binding motifs in the talin head control β3-integrin clustering. J. Cell Biol. 2009, 187, 715–731. [Google Scholar] [CrossRef] [Green Version]
- Janmey, P.A.; Bucki, R.; Radhakrishnan, R. Regulation of actin assembly by PI(4,5)P2 and other inositol phospholipids: An update on possible mechanisms. Biochem. Biophys. Res. Commun. 2018, 506, 307–314. [Google Scholar]
- Reversi, A.; Loeser, E.; Subramanian, D.; Schultz, C.; De Renzis, S. Plasma membrane phosphoinositide balance regulates cell shape during Drosophila embryo morphogenesis. J. Cell Biol. 2014, 205, 395–408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Viaud, J.; Mansour, R.; Antkowiak, A.; Mujalli, A.; Valet, C.; Chicanne, G.; Xuereb, J.M.; Terrisse, A.D.; Séverin, S.; Gratacap, M.P.; et al. Phosphoinositides: Important lipids in the coordination of cell dynamics. Biochimie 2016, 125, 250–258. [Google Scholar] [CrossRef] [PubMed]
- Goud, B.; Picas, L.; Gaits-Iacovoni, F. The emerging role of phosphoinositide clustering in intracellular trafficking and signal transduction. F1000Research 2016, 5. [Google Scholar] [CrossRef] [Green Version]
- Rameh, L.E.; Tolias, K.F.; Duckworth, B.C.; Cantley, L.C. A new pathway for synthesis of phosphatidylinositol-4,5-bisphosphate. Nature 1997, 390, 192–196. [Google Scholar] [CrossRef]
- Liu, C.; Deb, S.; Ferreira, V.S.; Xu, E.; Baumgart, T. Kinetics of PTEN-mediated PI(3,4,5)P3 hydrolysis on solid supported membranes. PLoS ONE 2018, 13, e0192667. [Google Scholar] [CrossRef] [Green Version]
- Thillai, K.; Lam, H.; Sarker, D.; Wells, C.M. Deciphering the link between PI3K and PAK: An opportunity to target key pathways in pancreatic cancer? Oncotarget 2017, 8, 14173–14191. [Google Scholar] [CrossRef] [Green Version]
- Vasudevan, K.M.; Barbie, D.A.; Davies, M.A.; Rabinovsky, R.; McNear, C.J.; Kim, J.J.; Hennessy, B.T.; Tseng, H.; Pochanard, P.; Kim, S.Y.; et al. AKT-independent signaling downstream of oncogenic PIK3CA mutations in human cancer. Cancer Cell 2009, 16, 21–32. [Google Scholar] [CrossRef] [Green Version]
- Balla, T. Phosphoinositides: Tiny lipids with giant impact on cell regulation. Physiol. Rev. 2013, 93, 1019–1137. [Google Scholar] [CrossRef]
- Goud, B.; Liu, S.; Storrie, B. Rab proteins as major determinants of the Golgi complex structure. Small GTPases 2018, 9, 66–75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heinrich, M.; Tian, A.; Esposito, C.; Baumgart, T. Dynamic sorting of lipids and proteins in membrane tubes with a moving phase boundary. Proc. Natl. Acad. Sci. USA 2010, 107, 7208–7213. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.L. Complex roles of PIP2 in the regulation of ion channels and transporters. Am. J. Physiol. Ren. Physiol. 2007, 293, 1761–1765. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez-Menchaca, A.A.; Adney, S.K.; Tang, Q.-Y.; Meng, X.-Y.; Rosenhouse-Dantsker, A.; Cui, M.; Logothetis, D.E.; Designed, D.E.L.; Performed, M.C. PIP 2 controls voltage-sensor movement and pore opening of Kv channels through the S4-S5 linker. Proc. Natl. Acad. Sci. USA 2012, 109, E2399–E2408. [Google Scholar] [CrossRef] [Green Version]
- Zaydman, M.A.; Silva, J.R.; Delaloye, K.; Li, Y.; Liang, H.; Larsson, H.P.; Shi, J.; Cui, J. Kv7.1 ion channels require a lipid to couple voltage sensing to pore opening. Proc. Natl. Acad. Sci. USA 2013, 110, 13180–13185. [Google Scholar] [CrossRef] [Green Version]
- Erle, D.J.; Sheppard, D. The cell biology of asthma. J. Cell Biol. 2014, 205, 621–631. [Google Scholar] [CrossRef] [Green Version]
- Mandal, K.; Wang, I.; Vitiello, E.; Orellana, L.A.C.; Balland, M. Cell dipole behaviour revealed by ECM sub-cellular geometry. Nat. Commun. 2014, 5. [Google Scholar] [CrossRef] [Green Version]
- Guet, D.; Mandal, K.; Pinot, M.; Hoffmann, J.; Abidine, Y.; Sigaut, W.; Bardin, S.; Schauer, K.; Goud, B.; Manneville, J.B. Mechanical role of actin dynamics in the rheology of the Golgi complex and in Golgi-associated trafficking events. Curr. Biol. 2014, 24, 1700–1711. [Google Scholar] [CrossRef] [Green Version]
- Mizuno, D.; Tardin, C.; Schmidt, C.F.; Mackintosh, F.C. Nonequilibrium mechanics of active cytoskeletal networks. Science 2007, 315, 370–373. [Google Scholar] [CrossRef] [PubMed]
- Mandal, K.; Balland, M.; Bureau, L. Thermoresponsive micropatterned substrates for single cell studies. PLoS ONE 2012, 7, e37548. [Google Scholar] [CrossRef] [Green Version]
- Mandal, K.; Asnacios, A.; Goud, B.; Manneville, J.-B. Mapping intracellular mechanics on micropatterned substrates. Proc. Natl. Acad. Sci. USA 2016, 113, E7159–E7168. [Google Scholar] [CrossRef] [Green Version]
- Honigmann, A.; Van Den Bogaart, G.; Iraheta, E.; Risselada, H.J.; Milovanovic, D.; Mueller, V.; Müllar, S.; Diederichsen, U.; Fasshauer, D.; Grubmüller, H.; et al. Phosphatidylinositol 4,5-bisphosphate clusters act as molecular beacons for vesicle recruitment. Nat. Struct. Mol. Biol. 2013, 20, 679–686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, H.L.; Janmey, P.A. Phosphoinositide Regulation of the Actin Cytoskeleton. Annu. Rev. Physiol. 2003, 65, 761–789. [Google Scholar] [CrossRef] [PubMed]
- Pogoda, K.; Bucki, R.; Byfield, F.J.; Cruz, K.; Lee, T.; Marcinkiewicz, C.; Janmey, P.A. Soft Substrates Containing Hyaluronan Mimic the Effects of Increased Stiffness on Morphology, Motility, and Proliferation of Glioma Cells. Biomacromolecules 2017, 18, 3040–3051. [Google Scholar] [CrossRef] [PubMed]
- Gowrishankar, K.; Ghosh, S.; Saha, S.; Mayor, S.; Rao, M. Active Remodeling of Cortical Actin Regulates Spatiotemporal Organization of Cell Surface Molecules. Cell 2012, 149, 1353–1367. [Google Scholar] [CrossRef] [Green Version]
- Mandal, K.; Raz-Ben Aroush, D.; Graber, Z.T.; Wu, B.; Park, C.Y.; Fredberg, J.J.; Guo, W.; Baumgart, T.; Janmey, P.A. Soft Hyaluronic Gels Promote Cell Spreading, Stress Fibers, Focal Adhesion, and Membrane Tension by Phosphoinositide Signaling, Not Traction Force. ACS Nano 2018, acsnano.8b05286. [Google Scholar] [CrossRef]
- Yamamoto, M.; Hilgemann, D.H.; Feng, S.; Bito, H.; Ishihara, H.; Shibasaki, Y.; Yin, H.L. Phosphatidylinositol 4,5-bisphosphate induces actin stress-fiber formation and inhibits membrane ruffling in CV1 cells. J. Cell Biol. 2001, 152, 867–876. [Google Scholar] [CrossRef] [Green Version]
- Lemmon, M.A. Membrane recognition by phospholipid-binding domains. Nat. Rev. Mol. Cell Biol. 2008, 9, 99–111. [Google Scholar] [CrossRef]
- Levine, T.P.; Munro, S. The pleckstrin homology domain of oxysterol-binding protein recognises a determinant specific to Golgi membranes. Curr. Biol. 1998, 8, 729–739. [Google Scholar] [CrossRef] [Green Version]
- Senju, Y.; Kalimeri, M.; Koskela, E.V.; Somerharju, P.; Zhao, H.; Vattulainen, I.; Lappalainen, P. Mechanistic principles underlying regulation of the actin cytoskeleton by phosphoinositides. Proc. Natl. Acad. Sci. USA 2017, 114, 8977–8986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bugyi, B.; Carlier, M.-F. Control of Actin Filament Treadmilling in Cell Motility. Ann. Rev. Biophys. 2010, 39, 449–470. [Google Scholar] [CrossRef]
- Bernstein, B.W.; Bamburg, J.R. ADF/Cofilin: A functional node in cell biology. Trends Cell Biol. 2010, 20, 187–195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakisaka, T.; Itoh, T.; Miura, K.; Takenawa, T. Phosphatidylinositol 4,5-bisphosphate phosphatase regulates the rearrangement of actin filaments. Mol. Cell. Biol. 1997, 17, 3841–3849. [Google Scholar] [CrossRef] [Green Version]
- Zhao, H.; Hakala, M.; Lappalainen, P. ADF/cofilin binds phosphoinositides in a multivalent manner to act as a PIP(2)-density sensor. Biophys. J. 2010, 98, 2327–2336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leyman, S.; Sidani, M.; Ritsma, L.; Waterschoot, D.; Eddy, R.; Dewitte, D.; Debeir, O.; Decaestecker, C.; Vandekerckhove, J.; Van Rheenen, J.; et al. Unbalancing the phosphatidylinositol-4,5-bisphosphate-cofilin interaction impairs cell steering. Mol. Biol. Cell 2009, 20, 4509–4523. [Google Scholar] [CrossRef] [Green Version]
- Sumi, T.; Matsumoto, K.; Takai, Y.; Nakamura, T. Cofilin phosphorylation and actin cytoskeletal dynamics regulated by Rho- and Cdc42-activated LIM-kinase 2. J. Cell Biol. 1999, 1519–1532. [Google Scholar] [CrossRef] [Green Version]
- Croisé, P.; Eestay-Aahumada, C.; Gasman, S.; Ory, S. Rho GTPases, phosphoinositides, and actin: A tripartite framework for efficient vesicular trafficking. Small GTPases 2014, 5. [Google Scholar] [CrossRef] [Green Version]
- Prunier, C.; Prudent, R.; Kapur, R.; Sadoul, K.; Lafanechère, L. LIM kinases: Cofilin and beyond. Oncotarget 2017, 8, 41749–41763. [Google Scholar] [CrossRef] [Green Version]
- Van Rheenen, J.; Song, X.; Van Roosmalen, W.; Cammer, M.; Chen, X.; DesMarais, V.; Yip, S.C.; Backer, J.M.; Eddy, R.J.; Condeelis, J.S. EGF-induced PIP2 hydrolysis releases and activates cofilin locally in carcinoma cells. J. Cell Biol. 2007, 179, 1247–1259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tuominen, E.K.J.; Holopainen, J.M.; Chen, J.; Prestwich, G.D.; Bachiller, P.R.; Kinnunen, P.K.J.; Janmey, P.A. Fluorescent phosphoinositide derivatives reveal specific binding of gelsolin and other actin regulatory proteins to mixed lipid bilayers. Eur. J. Biochem. 1999, 263, 85–92. [Google Scholar] [CrossRef] [Green Version]
- Janmey, P.A.; Stossel, T.P. Modulation of gelsolin function by phosphatidylinositol 4,5-Bisphosphate. Nature 1987, 325, 362–364. [Google Scholar] [CrossRef] [PubMed]
- Fatunmbi, O.; Bradley, R.P.; Kandy, S.K.; Bucki, R.; Janmey, P.A.; Radhakrishnan, R. A multiscale biophysical model for the recruitment of actin nucleating proteins at the membrane interface. Soft Matter 2020, 16, 4941–4954. [Google Scholar] [CrossRef]
- Wang, Y.H.; Bucki, R.; Janmey, P.A. Cholesterol-Dependent Phase-Demixing in Lipid Bilayers as a Switch for the Activity of the Phosphoinositide-Binding Cytoskeletal Protein Gelsolin. Biochemistry 2016, 55, 3361–3369. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Cao, L.; Wang, C.; Gigant, B.; Knossow, M. Kinesin, 30 years later: Recent insights from structural studies. Protein Sci. 2015, 24, 1047–1056. [Google Scholar] [CrossRef] [Green Version]
- Szatmári, D.; Xue, B.; Kannan, B.; Burtnick, L.D.; Bugyi, B.; Nyitrai, M.; Robinson, R.C. ATP competes with PIP2 for binding to gelsolin. PLoS ONE 2018, 13, e0201826. [Google Scholar] [CrossRef]
- Bucki, R.; Pastore, J.J.; Randhawa, P.; Vegners, R.; Weiner, D.J.; Janmey, P.A. Antibacterial Activities of Rhodamine B-Conjugated Gelsolin-Derived Peptides Compared to Those of the Antimicrobial Peptides Cathelicidin LL37, Magainin II, and Melittin. Antimicrob. Agents Chemother. 2004, 48, 1526–1533. [Google Scholar] [CrossRef] [Green Version]
- Bucki, R.; Niemirowicz-Laskowska, K.; Deptuła, P.; Wilczewska, A.Z.; Misiak, P.; Durnaś, B.; Fiedoruk, K.; Piktel, E.; Mystkowska, J.; Janmey, P.A. Susceptibility of microbial cells to the modified PIP2-binding sequence of gelsolin anchored on the surface of magnetic nanoparticles. J. Nanobiotechnology 2019, 17, 81. [Google Scholar] [CrossRef] [Green Version]
- Bucki, R.; Wang, Y.H.; Yang, C.; Kandy, S.K.; Fatunmbi, O.; Bradley, R.; Pogoda, K.; Svitkina, T.; Radhakrishnan, R.; Janmey, P.A. Lateral distribution of phosphatidylinositol 4,5-bisphosphate in membranes regulates formin- and ARP2/3-mediated actin nucleation. J. Biol. Chem. 2019, 294, 4704–4722. [Google Scholar] [CrossRef] [Green Version]
- Rohatgi, R.; Ma, L.; Miki, H.; Lopez, M.; Kirchhausen, T.; Takenawa, T.; Kirschner, M.W. The interaction between N-WASP and the Arp2/3 complex links Cdc42-dependent signals to actin assembly. Cell 1999, 97, 221–231. [Google Scholar] [CrossRef] [Green Version]
- Rohatgi, R.; Nollau, P.; Henry Ho, H.Y.; Kirschner, M.W.; Mayer, B.J. Nck and Phosphatidylinositol 4,5-Bisphosphate Synergistically Activate Actin Polymerization through the N-WASP-Arp2/3 Pathway. J. Biol. Chem. 2001, 276, 26448–26452. [Google Scholar] [CrossRef] [Green Version]
- Lassing, I.; Lindberg, U. Specific interaction between phosphatidylinositol 4,5-bisphosphate and profilactin. Nature 1985, 314, 472–474. [Google Scholar] [CrossRef]
- Bae, Y.H.; Ding, Z.; Das, T.; Wells, A.; Gertler, F.; Roy, P. Profilin1 regulates PI(3,4)P2 and lamellipodin accumulation at the leading edge thus influencing motility of MDA-MB-231 cells. Proc. Natl. Acad. Sci. USA 2010, 107, 21547–21552. [Google Scholar] [CrossRef] [Green Version]
- Goldschmidt-Clermont, P.J.; Machesky, L.M.; Baldassare, J.J.; Pollard, T.D. The actin-binding protein profilin binds to PIP2 and inhibits its hydrolysis by phospholipase C. Science 1990, 247, 1575–1578. [Google Scholar] [CrossRef]
- Goldschmidt-Clermont, P.J.; Furman, M.I.; Wachsstock, D.; Safer, D.; Nachmias, V.T.; Pollard, T.D. The control of actin nucleotide ex- change by thymosin beta 4 and profilin. A potential regulatory mechanism for actin polymerization in cells. Mol. Biol. Cell 1992, 3, 1015–1024. [Google Scholar] [CrossRef] [Green Version]
- Kovar, D.R.; Kuhn, J.R.; Tichy, A.L.; Pollard, T.D. The fission yeast cytokinesis formin Cdc12p is a barbed end actin filament capping protein gated by profilin. J. Cell Biol. 2003, 161, 875–887. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kowalski, J.R.; Egile, C.; Gil, S.; Snapper, S.B.; Li, R.; Thomas, S.M. Cortactin regulates cell migration through activation of N-WASP. J. Cell Sci. 2005, 118, 79–87. [Google Scholar] [CrossRef] [Green Version]
- Schafer, D.A.; Weed, S.A.; Binns, D.; Karginov, A.V.; Parsons, J.T.; Cooper, J.A. Dynamin2 and cortactin regulate actin assembly and filament organization. Curr. Biol. 2002, 12, 1852–1857. [Google Scholar] [CrossRef] [Green Version]
- Ribeiro, E.D.A.; Pinotsis, N.; Ghisleni, A.; Salmazo, A.; Konarev, P.V.; Kostan, J.; Sjöblom, B.; Schreiner, C.; Polyansky, A.A.; Gkougkoulia, E.A.; et al. The structure and regulation of human muscle α-Actinin. Cell 2014, 159, 1447–1460. [Google Scholar] [CrossRef] [Green Version]
- Pavalko, F.M.; Otey, C.A.; Simon, K.O.; Burridge, K. α-Actinin: A direct link between actin and integrins. Biochem. Soc. Trans. 1991, 19, 1065–1069. [Google Scholar] [CrossRef] [Green Version]
- Fukami, K.; Sawada, N.; Endo, T.; Takenawa, T. Identification of a Phosphatidylinositol 4,5-Bisphosphate-binding Site in Chicken Skeletal Muscle α-Actinin. J. Biol. Chem. 1996, 271, 2646–2650. [Google Scholar] [CrossRef] [Green Version]
- Sul, D.; Baron, C.B.; Broome, R.; Coburn, R.F. Smooth muscle length-dependent PI(4,5)P2 synthesis and paxillin tyrosine phosphorylation. Am. J. Physiol. Cell Physiol. 2001, 281, 300–310. [Google Scholar] [CrossRef] [Green Version]
- Kallikourdis, M.; Trovato, A.E.; Roselli, G.; Muscolini, M.; Porciello, N.; Tuosto, L.; Viola, A. Phosphatidylinositol 4-Phosphate 5-Kinase β Controls Recruitment of Lipid Rafts into the Immunological Synapse. J. Immunol. 2016, 196, 1955–1963. [Google Scholar] [CrossRef] [Green Version]
- Shabardina, V.; Kramer, C.; Gerdes, B.; Braunger, J.; Cordes, A.; Schäfer, J.; Mey, I.; Grill, D.; Gerke, V.; Steinem, C. Mode of Ezrin-Membrane Interaction as a Function of PIP2 Binding and Pseudophosphorylation. Biophys. J. 2016, 110, 2710–2719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jayasundar, J.J.; Ju, J.H.; He, L.; Liu, D.; Meilleur, F.; Zhao, J.; Callaway, D.J.E.; Bu, Z. Open conformation of ezrin bound to phosphatidylinositol 4,5-bisphosphate and to F-actin revealed by neutron scattering. J. Biol. Chem. 2012, 287, 37119–37133. [Google Scholar] [CrossRef] [Green Version]
- Feng, J.; Mertz, B. Novel Phosphotidylinositol 4,5-Bisphosphate Binding Sites on Focal Adhesion Kinase. PLoS ONE 2015, 10, e0132833. [Google Scholar] [CrossRef] [Green Version]
- Goñi, G.M.; Epifano, C.; Boskovic, J.; Camacho-Artacho, M.; Zhou, J.; Bronowska, A.; Martín, M.T.; Eck, M.J.; Kremer, L.; Gräter, F.; et al. Phosphatidylinositol 4,5-bisphosphate triggers activation of focal adhesion kinase by inducing clustering and conformational changes. Proc. Natl. Acad. Sci. USA 2014, 111, E3177–E3186. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Bronowska, A.; Le Coq, J.; Lietha, D.; Gräter, F. Allosteric regulation of focal adhesion kinase by PIP2 and ATP. Biophys. J. 2015, 108, 698–705. [Google Scholar] [CrossRef] [Green Version]
- Mcnamee, H.P.; Liley, H.G.; Ingber, D.E. Integrin-Dependent Control of Inositol Lipid Synthesis in Vascular Endothelial Cells and Smooth Muscle Cells. Exp. Cell Res. 1996, 224, 116–122. [Google Scholar] [CrossRef] [Green Version]
- Borowsky, M.L.; Hynes, R.O. Layilin, a novel talin-binding transmembrane protein homologous with C-type lectins, is localized in membrane ruffles. J. Cell Biol. 1998, 143, 429–442. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Zhao, L.; Suzuki, A.; Lian, L.; Min, S.H.; Wang, Z.; Litvinov, R.I.; Stalker, T.J.; Yago, T.; Klopocki, A.G.; et al. Platelets lacking PIP5KIγ have normal integrin activation but impaired cytoskeletal-membrane integrity and adhesion. Blood 2013, 121, 2743–2752. [Google Scholar] [CrossRef] [Green Version]
- Das, M.; Ithychanda, S.; Qin, J.; Plow, E.F. Mechanisms of talin-dependent integrin signaling and crosstalk. Biochim. Biophys. Acta 2014, 1838, 579–588. [Google Scholar] [CrossRef] [Green Version]
- Moore, D.T.; Nygren, P.; Jo, H.; Boesze-Battaglia, K.; Bennett, J.S.; DeGrado, W.F. Affinity of talin-1 for the β3-integrin cytosolic domain is modulated by its phospholipid bilayer environment. Proc. Natl. Acad. Sci. USA 2012, 109, 793–798. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elliott, P.R.; Goult, B.T.; Kopp, P.M.; Bate, N.; Grossmann, J.G.; Roberts, G.C.K.; Critchley, D.R.; Barsukov, I.L. The Structure of the Talin Head Reveals a Novel Extended Conformation of the FERM Domain. Structure 2010, 18, 1289–1299. [Google Scholar] [CrossRef] [Green Version]
- Thompson, P.M.; Ramachandran, S.; Case, L.B.; Tolbert, C.E.; Tandon, A.; Pershad, M.; Dokholyan, N.V.; Waterman, C.M.; Campbell, S.L. A Structural Model for Vinculin Insertion into PIP2-Containing Membranes and the Effect of Insertion on Vinculin Activation and Localization. Structure 2017, 25, 264–275. [Google Scholar] [CrossRef] [Green Version]
- Chandrasekar, I.; Stradal, T.E.B.; Holt, M.R.; Entschladen, F.; Jockusch, B.M.; Ziegler, W.H. Vinculin acts as a sensor in lipid regulation of adhesion-site turnover. J. Cell Sci. 2005, 118, 1461–1472. [Google Scholar] [CrossRef] [Green Version]
- Di Paolo, G.; De Camilli, P. Phosphoinositides in cell regulation and membrane dynamics. Nature 2006, 443, 651–657. [Google Scholar] [CrossRef]
- Wang, J.; Richards, D.A. Segregation of PIP2 and PIP3 into distinct nanoscale regions within the plasma membrane. Biol. Open 2012, 1, 857–862. [Google Scholar] [CrossRef] [Green Version]
- Van Den Bogaart, G.; Meyenberg, K.; Risselada, H.J.; Amin, H.; Willig, K.I.; Hubrich, B.E.; Dier, M.; Hell, S.W.; Grubmüller, H.; Diederichsen, U.; et al. Membrane protein sequestering by ionic protein-lipid interactions. Nature 2011, 479, 552–555. [Google Scholar] [CrossRef] [PubMed]
- McLaughlin, S. The Electrostatic Properties of Membranes. Ann. Rev. Biophys. Biophys. Chem. 1989, 18, 113–136. [Google Scholar] [CrossRef]
- Gambhir, A.; Hangyás-Mihályné, G.; Zaitseva, I.; Cafiso, D.S.; Wang, J.; Murray, D.; Pentyala, S.N.; Smith, S.O.; McLaughlin, S. Electrostatic sequestration of PIP2 on phospholipid membranes by basic/aromatic regions of proteins. Biophys. J. 2004, 86, 2188–2207. [Google Scholar] [CrossRef] [Green Version]
- Cevc, G.; Svetina, S.; Žekš, B. Electrostatic potential of bilayer lipid membranes with the structural surface charge smeared perpendicular to the membrane-solution interface. An extension of the Gouy-Chapman diffuse double layer theory. J. Phys. Chem. 1981, 85, 1762–1767. [Google Scholar] [CrossRef]
- Shapovalov, V.L.; Brezesinski, G. Breakdown of the Gouy—Chapman model for highly charged Langmuir monolayers: Counterion size effect. J. Phys. Chem. B 2006, 110, 10032–10040. [Google Scholar] [CrossRef]
- Li, Z.; Venable, R.M.; Rogers, L.A.; Murray, D.; Pastor, R.W. Molecular dynamics simulations of PIP2 and PIP3 in lipid bilayers: Determination of ring orientation, and the effects of surface roughness on a Poisson-Boltzmann description. Biophys. J. 2009, 97, 155–163. [Google Scholar] [CrossRef] [Green Version]
- Slochower, D.R.; Huwe, P.J.; Radhakrishnan, R.; Janmey, P.A. Quantum and all-atom molecular dynamics simulations of protonation and divalent ion binding to phosphatidylinositol 4,5-bisphosphate (PIP2). J. Phys. Chem. B 2013, 117, 8322–8329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McLaughlin, S.; Wang, J.; Gambhir, A.; Murray, D. PIP 2 and Proteins: Interactions, Organization, and Information Flow. Annu. Rev. Biophys. Biomol. Struct. 2002, 31, 151–175. [Google Scholar] [CrossRef] [Green Version]
- Levental, I.; Christian, D.A.; Wang, Y.H.; Madara, J.J.; Discher, D.E.; Janmey, P.A. Calcium-dependent lateral organization in phosphatidylinositol 4,5-bisphosphate (PIP2)- and cholesterol-containing monolayers. Biochemistry 2009, 48, 8241–8248. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.H.; Collins, A.; Guo, L.; Smith-Dupont, K.B.; Gai, F.; Svitkina, T.; Janmey, P.A. Divalent cation-induced cluster formation by polyphosphoinositides in model membranes. J. Am. Chem. Soc. 2012, 134, 3387–3395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bradley, R.P.; Slochower, D.R.; Janmey, P.A.; Radhakrishnan, R. Divalent cations bind to phosphoinositides to induce ion and isomer specific propensities for nano-cluster initiation in bilayer membranes. R. Soc. Open Sci. 2020, 7, 192208. [Google Scholar] [CrossRef]
- Wen, Y.; Vogt, V.M.; Feigenson, G.W. Multivalent Cation-Bridged PI(4,5)P2 Clusters Form at Very Low Concentrations. Biophys. J. 2018, 114, 2630–2639. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balla, T.; Bondeva, T.; Várnai, P. How accurately can we image inositol lipids in living cells? Trends Pharmacol. Sci. 2000, 21, 238–241. [Google Scholar] [CrossRef]
- Sorre, B.; Callan-Jones, A.; Manneville, J.B.; Nassoy, P.; Joanny, J.F.; Prost, J.; Goud, B.; Bassereau, P. Curvature-driven lipid sorting needs proximity to a demixing point and is aided by proteins. Proc. Natl. Acad. Sci. USA 2009, 106, 5622–5626. [Google Scholar] [CrossRef] [Green Version]
- Roux, A.; Koster, G.; Lenz, M.; Sorre, B.; Manneville, J.B.; Nassoy, P.; Bassereau, P. Membrane curvature controls dynamin polymerization. Proc. Natl. Acad. Sci. USA 2010, 107, 4141–4146. [Google Scholar] [CrossRef] [Green Version]
- Lin, X.; Wang, H.; Lou, Z.; Cao, M.; Zhang, Z.; Gu, N. Roles of PIP 2 in the membrane binding of MIM I- BAR: Insights from molecular dynamics simulations. FEBS Lett. 2018, 592, 2533–2542. [Google Scholar] [CrossRef] [Green Version]
- Simunovic, M.; Manneville, J.-B.; Renard, H.-F.; Evergren, E.; Raghunathan, K.; Bhatia, D.; Kenworthy, A.K.; Voth, G.A.; Prost, J.; McMahon, H.T.; et al. Friction Mediates Scission of Tubular Membranes Scaffolded by BAR Proteins. Cell 2017, 170, 172–184.e11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, T.; Baumgart, T. BIN1 Membrane Curvature Sensing and Generation Show Autoinhibition Regulated by Downstream Ligands and PI(4,5)P 2. Biochemistry 2014. [Google Scholar] [CrossRef] [Green Version]
- Wu, T.; Shi, Z.; Baumgart, T. Mutations in BIN1 Associated with Centronuclear Myopathy Disrupt Membrane Remodeling by Affecting Protein Density and Oligomerization. PLoS ONE 2014, 9, e93060. [Google Scholar] [CrossRef]
- Nicot, A.S.; Toussaint, A.; Tosch, V.; Kretz, C.; Wallgren-Pettersson, C.; Iwarsson, E.; Kingston, H.; Garnier, J.M.; Biancalana, V.; Oldfors, A.; et al. Mutations in amphiphysin 2 (BIN1) disrupt interaction with dynamin 2 and cause autosomal recessive centronuclear myopathy. Nat. Genet. 2007, 39, 1134–1139. [Google Scholar] [CrossRef]
- Brown, D.A.; Hughes, S.A.; Marsh, S.J.; Tinker, A. Regulation of M(Kv7.2/7.3) channels in neurons by PIP 2 and products of PIP 2 hydrolysis: Significance for receptor-mediated inhibition. J. Physiol. 2007, 582, 917–925. [Google Scholar] [CrossRef]
- Ebi, H.; Costa, C.; Faber, A.C.; Nishtala, M.; Kotani, H.; Juric, D.; Della Pelle, P.; Song, Y.; Yano, S.; Mino-Kenudson, M.; et al. PI3K regulates MEK/ERK signaling in breast cancer via the Rac-GEF, P-Rex1. Proc. Natl. Acad. Sci. USA 2013, 110, 21124–21129. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Prakash, P.; Gorfe, A.A.; Hancock, J.F. Ras and the Plasma Membrane: A Complicated Relationship. Cold Spring Harb. Perspect. Med. 2018, 8. [Google Scholar] [CrossRef] [Green Version]
- Falkenburger, B.H.; Jensen, J.B.; Dickson, E.J.; Suh, B.C.; Hille, B. Phosphoinositides: Lipid regulators of membrane proteins. J. Physiol. 2010, 588, 3179–3185. [Google Scholar] [CrossRef]
- Szentpetery, Z.; Várnai, P.; Balla, T. Acute manipulation of Golgi phosphoinositides to assess their importance in cellular trafficking and signaling. Proc. Natl. Acad. Sci. USA 2010, 107, 8225–8230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Evans, P.R.; Owen, D.J. Endocytosis and vesicle trafficking. Curr. Opin. Struct. Biol. 2002, 12, 814–821. [Google Scholar] [CrossRef]
- Posor, Y.; Eichhorn-Gruenig, M.; Puchkov, D.; Schöneberg, J.; Ullrich, A.; Lampe, A.; Müller, R.; Zarbakhsh, S.; Gulluni, F.; Hirsch, E.; et al. Spatiotemporal control of endocytosis by phosphatidylinositol-3,4- bisphosphate. Nature 2013, 499, 233–237. [Google Scholar] [CrossRef]
- Goud, B. Why does endocytosis in single cells care which side up? Bioarchitecture 2014, 4, 62–67. [Google Scholar]
- Jost, M.; Simpson, F.; Kavran, J.M.; Lemmon, M.A.; Schmid, S.L. Phosphatidylinositol-4,5-bisphosphate is required for endocytic coated vesicle formation. Curr. Biol. 1998, 8, 1399–1404. [Google Scholar] [CrossRef] [Green Version]
- Czech, M.P. PIP2 and PIP3: Complex roles at the cell surface. Cell 2000, 100, 603–606. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; Hedman, A.C.; Tan, X.; Schill, N.J.; Anderson, R.A. Endosomal Type Iγ PIP 5-Kinase Controls EGF Receptor Lysosomal Sorting. Dev. Cell 2013, 25, 144–155. [Google Scholar] [CrossRef] [Green Version]
- Gullapalli, A.; Garrett, T.A.; Paing, M.M.; Griffin, C.T.; Yang, Y.; Trejo, J.A. A Role for Sorting Nexin 2 in Epidermal Growth Factor Receptor Down-regulation: Evidence for Distinct Functions of Sorting Nexin 1 and 2 in Protein Trafficking. Mol. Biol. Cell 2004, 15, 2143–2155. [Google Scholar] [CrossRef] [Green Version]
- Merino-Trigo, A.; Kerr, M.C.; Houghton, F.; Lindberg, A.; Mitchell, C.; Teasdale, R.D.; Gleeson, P.A. Sorting nexin 5 is localized to a subdomain of the early endosomes and is recruited to the plasma membrane following EGF stimulation. J. Cell Sci. 2004, 117, 6413–6424. [Google Scholar] [CrossRef] [Green Version]
- Cheung, M.; Sharma, A.; Madhunapantula, S.R.V.; Robertson, G.P. Akt3 and mutant V600EB-Raf cooperate to promote early melanoma development. Cancer Res. 2008, 68, 3429–3439. [Google Scholar] [CrossRef] [Green Version]
- Amblard, I.; Dupont, E.; Alves, I.; Miralves, J.; Queguiner, I.; Joliot, A. Bidirectional transfer of homeoprotein EN2 across the plasma membrane requires PIP2. J. Cell Sci. 2020, 133. [Google Scholar] [CrossRef]
- De Matteis, M.A.; Godi, A. PI-loting membrane traffic. Nat. Cell Biol. 2004, 6, 487–492. [Google Scholar] [CrossRef]
- De Craene, J.O.; Bertazzi, D.L.; Bär, S.; Friant, S. Phosphoinositides, major actors in membrane trafficking and lipid signaling pathways. Int. J. Mol. Sci. 2017, 18, 634. [Google Scholar] [CrossRef]
- Krauß, M.; Haucke, V. Phosphoinositide-metabolizing enzymes at the interface between membrane traffic and cell signalling. EMBO Rep. 2007, 8, 241–246. [Google Scholar] [CrossRef] [Green Version]
- Balla, T.; Várnai, P. Visualization of cellular phosphoinositide pools with GFP-fused protein-domains. Curr. Protoc. Cell Biol. 2009, 42, 24.4.1–24.4.27. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Du, X.N.; Jia, Q.Z.; Zhang, H.L. Binding of PLCδ1PH-GFP to Ptdlns(4,5)P2 prevents inhibition of phospholipase C-mediated hydrolysis of Ptdlns(4,5)P 2 by neomycin. Acta Pharmacol. Sin. 2005, 26, 1485–1491. [Google Scholar] [CrossRef] [Green Version]
- Poli, A.; Zaurito, A.E.; Abdul-Hamid, S.; Fiume, R.; Faenza, I.; Divecha, N. Phosphatidylinositol 5 phosphate (Pi5p): From behind the scenes to the front (nuclear) stage. Int. J. Mol. Sci. 2019, 20, 2080. [Google Scholar] [CrossRef] [Green Version]
- Gary, J.D.; Wurmser, A.E.; Bonangelino, C.J.; Weisman, L.S.; Emr, S.D. Fab1p is essential for PtdIns(3)P 5-kinase activity and the maintenance of vacuolar size and membrane homeostasis. J. Cell Biol. 1998, 143, 65–79. [Google Scholar] [CrossRef]
- Eugster, A.; Pécheur, E.I.; Michel, F.; Winsor, B.; Letourneur, F.; Friant, S. Ent5p is required with Ent3p and Vps27p for ubiquitin-dependent protein sorting into the multivesicular body. Mol. Biol. Cell 2004, 15, 3031–3041. [Google Scholar] [CrossRef]
- Chalhoub, N.; Baker, S.J. PTEN and the PI3-kinase pathway in cancer. Annu. Rev. Pathol. Mech. Dis. 2009, 4, 127–150. [Google Scholar] [CrossRef] [Green Version]
- Zimmerman, M.W.; Homanics, G.E.; Lazo, J.S. Targeted Deletion of the Metastasis-Associated Phosphatase Ptp4a3 (PRL-3) Suppresses Murine Colon Cancer. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Marshall, A.J. Phosphatidylinositol (3,4) bisphosphate-specific phosphatases and effector proteins: A distinct branch of PI3K signaling. Cell. Signal. 2015, 27, 1789–1798. [Google Scholar] [CrossRef]
- Cheung, S.M.S.; Kornelson, J.C.; Al-Alwan, M.; Marshall, A.J. Regulation of phosphoinositide 3-kinase signaling by oxidants: Hydrogen peroxide selectively enhances immunoreceptor-induced recruitment of phosphatidylinositol (3,4) bisphosphate-binding PH domain proteins. Cell. Signal. 2007, 19, 902–912. [Google Scholar] [CrossRef]
- Cunningham, C.C.; Vegners, R.; Bucki, R.; Funaki, M.; Korde, N.; Hartwig, J.H.; Stossel, T.P.; Janmey, P.A. Cell permeant polyphosphoinositide-binding peptides that block cell motility and actin assembly. J. Biol. Chem. 2001, 276, 43390–43399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choudhury, R.; Diao, A.; Zhang, F.; Eisenberg, E.; Saint-Pol, A.; Williams, C.; Konstantakopoulos, A.; Lucocq, J.; Johannes, L.; Rabouille, C.; et al. Lowe syndrome protein OCRL1 interacts with clathrin and regulates protein trafficking between endosomes and the trans-Golgi network. Mol. Biol. Cell 2005, 16, 3467–3479. [Google Scholar] [CrossRef] [Green Version]
- Vicinanza, M.; Di Campli, A.; Polishchuk, E.; Santoro, M.; Di Tullio, G.; Godi, A.; Levtchenko, E.; De Leo, M.G.; Polishchuk, R.; Sandoval, L.; et al. OCRL controls trafficking through early endosomes via PtdIns4,5P 2-dependent regulation of endosomal actin. EMBO J. 2011, 30, 4970–4985. [Google Scholar] [CrossRef] [PubMed]
- Cremona, O.; Di Paolo, G.; Wenk, M.R.; Lüthi, A.; Kim, W.T.; Takei, K.; Daniell, L.; Nemoto, Y.; Shears, S.B.; Flavell, R.A.; et al. Essential role of phosphoinositide metabolism in synaptic vesicle recycling. Cell 1999, 99, 179–188. [Google Scholar] [CrossRef] [Green Version]
- Mücksch, F.; Citir, M.; Lüchtenborg, C.; Glass, B.; Traynor-Kaplan, A.; Schultz, C.; Brügger, B.; Kräusslich, H.G. Quantification of phosphoinositides reveals strong enrichment of PIP2 in HIV-1 compared to producer cell membranes. Sci. Rep. 2019, 9. [Google Scholar] [CrossRef]
- Rocha-Perugini, V.; Gordon-Alonso, M.; Sánchez-Madrid, F. PIP2: Choreographer of actin-adaptor proteins in the HIV-1 dance. Trends Microbiol. 2014, 22, 379–388. [Google Scholar] [CrossRef] [Green Version]
- Arancio, O. PIP2: A new key player in Alzheimer’s disease. Cellscience 2008, 5, 44–47. [Google Scholar]
- Duciel, L.; Anezo, O.; Mandal, K.; Laurent, C.; Planque, N.; Coquelle, F.M.; Gentien, D.; Manneville, J.-B.; Saule, S. Protein tyrosine phosphatase 4A3 (PTP4A3/PRL-3) promotes the aggressiveness of human uveal melanoma through dephosphorylation of CRMP2. Sci. Rep. 2019, 9, 2990. [Google Scholar] [CrossRef]
- Mandal, K.; Gong, Z.; Rylander, A.; Shenoy, V.B.; Janmey, P. Opposite responses of normal hepatocytes and hepatocellular carcinoma cells to substrate viscoelasticity. Biomater. Sci. 2020, 8, 1316–1328. [Google Scholar] [CrossRef]
- Lan, T.; Pang, J.; Wu, Y.; Zhu, M.; Yao, X.; Wu, M.; Qian, H.; Zhang, Z.; Gao, J.; Chen, Y. Cross-linked hyaluronic acid gel inhibits metastasis and growth of gastric and hepatic cancer cells: In vitro and in vivo studies. Oncotarget 2016, 7, 65418–65428. [Google Scholar] [CrossRef]
- Guillou, H.; Lécureuil, C.; Anderson, K.E.; Suire, S.; Ferguson, G.J.; Ellson, C.D.; Gray, A.; Divecha, N.; Hawkins, P.T.; Stephens, L.R. Use of the GRP1 PH domain as a tool to measure the relative levels of PtdIns(3,4,5)P3 through a protein-lipid overlay approach. J. Lipid Res. 2007, 48, 726–732. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Shao, R.; Zhang, X.D.; Chen, C. Applications of nanotechnology for melanoma treatment, diagnosis, and theranostics. Int. J. Nanomed. 2013, 8, 2677–2688. [Google Scholar] [CrossRef] [Green Version]
- McParland, V.; Varsano, G.; Li, X.; Thornton, J.; Baby, J.; Aravind, A.; Meyer, C.; Pavic, K.; Rios, P.; Köhn, M. The metastasis-promoting phosphatase PRL-3 shows activity toward phosphoinositides. Biochemistry 2011, 50, 7579–7590. [Google Scholar] [CrossRef]
- Liu, J.; Gardel, M.L.; Kroy, K.; Frey, E.; Hoffman, B.D.; Crocker, J.C.; Bausch, A.R.; Weitz, D.A. Microrheology Probes Length Scale Dependent Rheology. Phys. Rev. Lett. 2005. [Google Scholar] [CrossRef] [Green Version]
- Daly, R.J. Cortactin signalling and dynamic actin networks. Biochem. J. 2004, 382, 13–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]



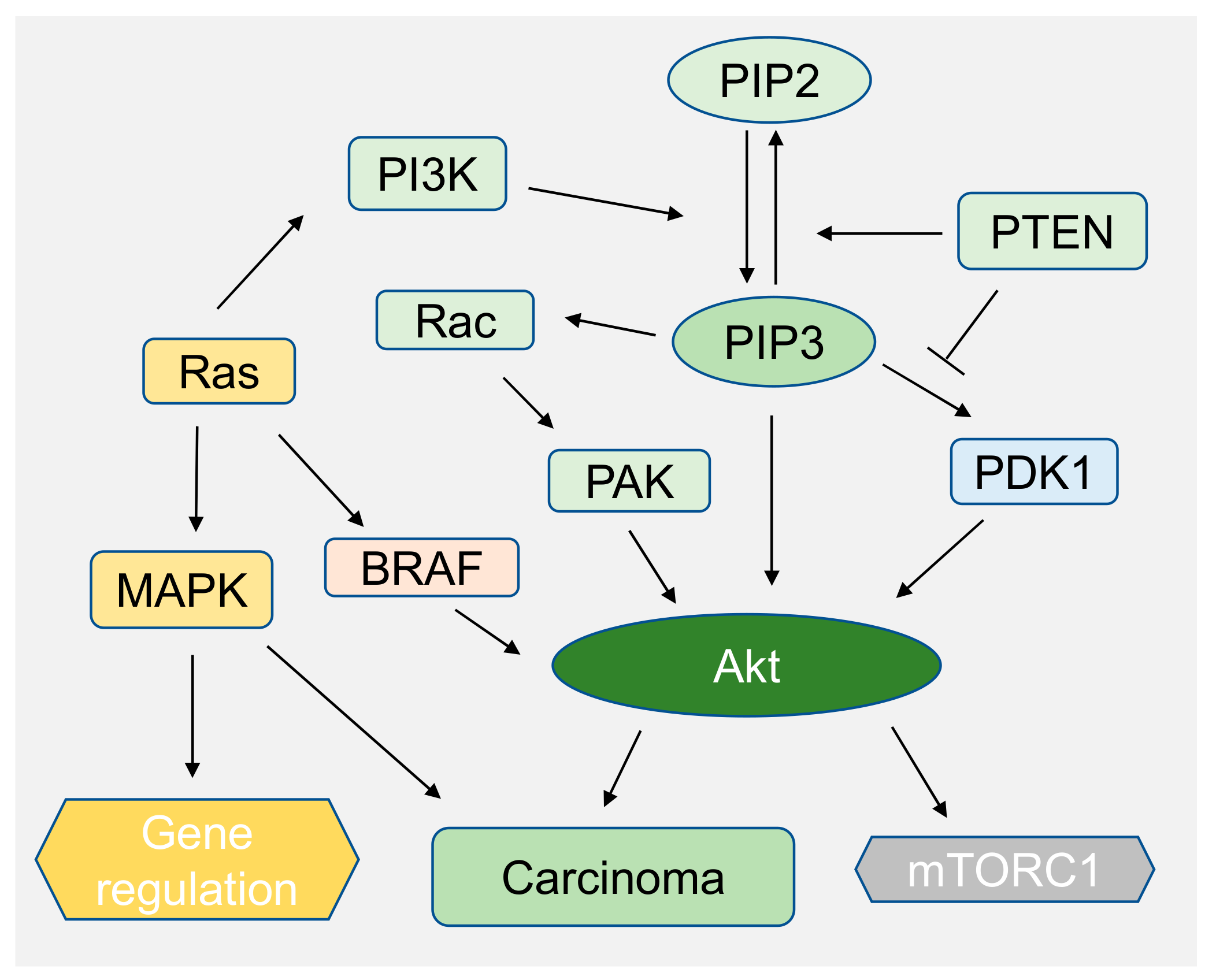
| Phospho-Inositides | Pathways/Functions | Enzymatic Activity | Disease Implication | References |
|---|---|---|---|---|
| PIP3, PI(4,5) P2, PI(3,5) P2 | PI3K-Akt | PI3K, PTEN 1,2 | Melanoma cancer, Cowden disease, pancreatic cancer, ovarian cancer. | [132] |
| PI(4,5) P2 | Endocytic trafficking pathways | OCRL, 5 phosphatases | Oculo-cerebro-renal syndrome of Lowe: renal Fancomi syndrome, glaucoma, cataracts, blindness, mental retardation. | [133,134] |
| PI(3,5) P2 | MTM1, PI4P, | Myopathy. | [2,121] | |
| PI(3,5) P2 | Fab1/PIKfyve kinase | Neuropathologies, Charcot-Marie tooth disease. | ||
| PI(4,5) P2, PI(3,5) P2 | Endocytic pathways | Synaptojanin1,2 | Bipolar disorder, Down syndrome, neuronal disorder. | [29,85,135] |
| PI(4,5) P2 | INPP4 | Asthma, nondegeneracy. | [23,130] | |
| PI(4,5) P2 | Actin reorganization | Human immunodeficiency virus-1 (HIV-1). | [136,137] | |
| PI(4,5) P2 | impairment of synaptic function | Amyloid-β peptide oligomers | Alzheimer’s disease | [138] |
| PI(3,4) P2. PIP3 | Akt/PKB | Cell survival and growth, cancer. | [16,130,131] | |
| PI(4,5) P2 | PRL-3 | Melanoma, colon cancer. | [131,136,137] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mandal, K. Review of PIP2 in Cellular Signaling, Functions and Diseases. Int. J. Mol. Sci. 2020, 21, 8342. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21218342
Mandal K. Review of PIP2 in Cellular Signaling, Functions and Diseases. International Journal of Molecular Sciences. 2020; 21(21):8342. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21218342
Chicago/Turabian StyleMandal, Kalpana. 2020. "Review of PIP2 in Cellular Signaling, Functions and Diseases" International Journal of Molecular Sciences 21, no. 21: 8342. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21218342




