Cannabinoid Type 1 Receptor is Undetectable in Rodent and Primate Cerebral Neural Stem Cells but Participates in Radial Neuronal Migration
Abstract
:1. Introduction
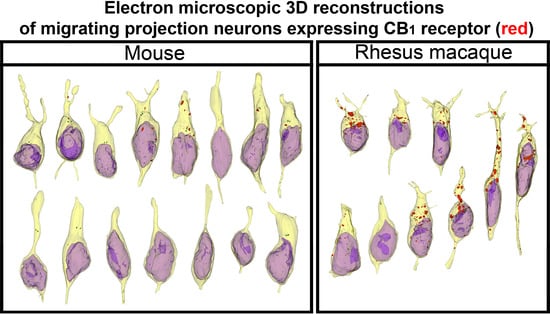
2. Results
3. Discussion
4. Materials and Methods
4.1. In Silico Quantification of Cannabinoid System Genes Expression in the Mouse Embryo Cerebral Cortex
4.2. Animals
4.3. IHC Labeling for Light and Electron Microscopy
4.4. Serial Ultrathin Sectioning and Electron Microscopic 3D Reconstruction
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| CB1R | Cannabinoid type 1 receptor |
| 3D | three-dimensional |
| VZ | ventricular zone |
| SVZ | subventricular zone |
| IZ | intermediate zone |
| CP | cortical plate |
| MZ | marginal zone |
| NSC | neural stem cell |
| IHC | immunohistochemical |
| E | embryonic day |
| CB1R−/− | CB1R-knock out |
| CB1R-L15 | antibodies generated against last 15 amino acids of CB1R |
| CB1R-L31 | antibodies generated against last 31 amino acids of CB1R |
| CB1R-NH | antibodies generated against amino-terminus of CB1R |
| DAB | 3,3′-diaminobenzidine-4HCl |
| DAB-Ni | Ni-intensified 3,3′-diaminobenzidine-4HCl |
References
- Geschwind, D.H.; Rakic, P. Cortical evolution: Judge the brain by its cover. Neuron 2013, 80, 633–647. [Google Scholar] [CrossRef] [Green Version]
- Rakic, P. Evolution of the neocortex: A perspective from developmental biology. Nat. Rev. Neurosci. 2009, 10, 724–735. [Google Scholar] [CrossRef]
- Rakic, P.; Ayoub, A.E.; Breunig, J.J.; Dominguez, M.H. Decision by division: Making cortical maps. Trends Neurosci. 2009, 32, 291–301. [Google Scholar] [CrossRef] [Green Version]
- Hippenmeyer, S. Molecular pathways controlling the sequential steps of cortical projection neuron migration. Adv. Exp. Med. Biol. 2014, 800, 1–24. [Google Scholar] [CrossRef]
- Fukuda, T.; Yanagi, S. Psychiatric behaviors associated with cytoskeletal defects in radial neuronal migration. Cell. Mol. Life Sci. 2017, 74, 3533–3552. [Google Scholar] [CrossRef]
- Kast, R.J.; Levitt, P. Precision in the development of neocortical architecture: From progenitors to cortical networks. Prog. Neurobiol. 2019, 175, 77–95. [Google Scholar] [CrossRef]
- Lasser, M.; Tiber, J.; Lowery, L.A. The Role of the Microtubule Cytoskeleton in Neurodevelopmental Disorders. Front. Cell. Neurosci. 2018, 12, 165. [Google Scholar] [CrossRef] [Green Version]
- Sidman, R.L.; Rakic, P. Neuronal migration, with special reference to developing human brain: A review. Brain Res. 1973, 62, 1–35. [Google Scholar] [CrossRef]
- Kosodo, Y.; Suetsugu, T.; Suda, M.; Mimori-Kiyosue, Y.; Toida, K.; Baba, S.A.; Kimura, A.; Matsuzaki, F. Regulation of interkinetic nuclear migration by cell cycle-coupled active and passive mechanisms in the developing brain. Embo J. 2011, 30, 1690–1704. [Google Scholar] [CrossRef]
- Joukov, V.; De Nicolo, A. The Centrosome and the Primary Cilium: The Yin and Yang of a Hybrid Organelle. Cells 2019, 8, 701. [Google Scholar] [CrossRef] [Green Version]
- Azimzadeh, J.; Marshall, W.F. Building the centriole. Curr. Biol. 2010, 20, R816–R825. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agircan, F.G.; Schiebel, E.; Mardin, B.R. Separate to operate: Control of centrosome positioning and separation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014, 369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsuzaki, F.; Shitamukai, A. Cell Division Modes and Cleavage Planes of Neural Progenitors during Mammalian Cortical Development. Cold Spring Harb. Perspect. Biol. 2015, 7, a015719. [Google Scholar] [CrossRef] [PubMed]
- Higginbotham, H.R.; Gleeson, J.G. The centrosome in neuronal development. Trends Neurosci. 2007, 30, 276–283. [Google Scholar] [CrossRef]
- Veland, I.R.; Lindbæk, L.; Christensen, S.T. Linking the Primary Cilium to Cell Migration in Tissue Repair and Brain Development. Bioscience 2014, 64, 1115–1125. [Google Scholar] [CrossRef]
- Mirvis, M.; Stearns, T.; James Nelson, W. Cilium structure, assembly, and disassembly regulated by the cytoskeleton. Biochem. J. 2018, 475, 2329–2353. [Google Scholar] [CrossRef] [Green Version]
- Arellano, J.I.; Guadiana, S.M.; Breunig, J.J.; Rakic, P.; Sarkisian, M.R. Development and distribution of neuronal cilia in mouse neocortex. J. Comp. Neurol. 2012, 520, 848–873. [Google Scholar] [CrossRef] [Green Version]
- Matsumoto, M.; Sawada, M.; Garcia-Gonzalez, D.; Herranz-Perez, V.; Ogino, T.; Bang Nguyen, H.; Quynh Thai, T.; Narita, K.; Kumamoto, N.; Ugawa, S.; et al. Dynamic Changes in Ultrastructure of the Primary Cilium in Migrating Neuroblasts in the Postnatal Brain. J. Neurosci. 2019, 39, 9967–9988. [Google Scholar] [CrossRef]
- Katona, I.; Freund, T.F. Multiple functions of endocannabinoid signaling in the brain. Annu. Rev. Neurosci. 2012, 35, 529–558. [Google Scholar] [CrossRef] [Green Version]
- Piomelli, D. A mighty (ochondrial) fight? Mol. Metab. 2014, 3, 345–346. [Google Scholar] [CrossRef]
- Hurd, Y.L.; Manzoni, O.J.; Pletnikov, M.V.; Lee, F.S.; Bhattacharyya, S.; Melis, M. Cannabis and the Developing Brain: Insights into Its Long-Lasting Effects. J. Neurosci. 2019, 39, 8250–8258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berghuis, P.; Rajnicek, A.M.; Morozov, Y.M.; Ross, R.A.; Mulder, J.; Urban, G.M.; Monory, K.; Marsicano, G.; Matteoli, M.; Canty, A.; et al. Hardwiring the brain: Endocannabinoids shape neuronal connectivity. Science 2007, 316, 1212–1216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keimpema, E.; Barabas, K.; Morozov, Y.M.; Tortoriello, G.; Torii, M.; Cameron, G.; Yanagawa, Y.; Watanabe, M.; Mackie, K.; Harkany, T. Differential subcellular recruitment of monoacylglycerol lipase generates spatial specificity of 2-arachidonoyl glycerol signaling during axonal pathfinding. J. Neurosci. 2010, 30, 13992–14007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morozov, Y.M.; Torii, M.; Rakic, P. Origin, early commitment, migratory routes, and destination of cannabinoid type 1 receptor-containing interneurons. Cereb. Cortex (N.Y. 1991) 2009, 19 (Suppl. 1), i78–i89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diaz-Alonso, J.; de Salas-Quiroga, A.; Paraiso-Luna, J.; Garcia-Rincon, D.; Garcez, P.P.; Parsons, M.; Andradas, C.; Sanchez, C.; Guillemot, F.; Guzman, M.; et al. Loss of Cannabinoid CB1 Receptors Induces Cortical Migration Malformations and Increases Seizure Susceptibility. Cereb. Cortex (N.Y. 1991) 2017, 27, 5303–5317. [Google Scholar] [CrossRef]
- Oudin, M.J.; Gajendra, S.; Williams, G.; Hobbs, C.; Lalli, G.; Doherty, P. Endocannabinoids regulate the migration of subventricular zone-derived neuroblasts in the postnatal brain. J. Neurosci. 2011, 31, 4000–4011. [Google Scholar] [CrossRef]
- Diaz-Alonso, J.; Guzman, M.; Galve-Roperh, I. Endocannabinoids via CB(1) receptors act as neurogenic niche cues during cortical development. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2012, 367, 3229–3241. [Google Scholar] [CrossRef] [Green Version]
- Galve-Roperh, I.; Chiurchiu, V.; Diaz-Alonso, J.; Bari, M.; Guzman, M.; Maccarrone, M. Cannabinoid receptor signaling in progenitor/stem cell proliferation and differentiation. Prog. Lipid Res. 2013, 52, 633–650. [Google Scholar] [CrossRef]
- Sagredo, O.; Palazuelos, J.; Gutierrez-Rodriguez, A.; Satta, V.; Galve-Roperh, I.; Martinez-Orgado, J. Cannabinoid signalling in the immature brain: Encephalopathies and neurodevelopmental disorders. Biochem. Pharmacol. 2018, 157, 85–96. [Google Scholar] [CrossRef]
- Aguado, T.; Monory, K.; Palazuelos, J.; Stella, N.; Cravatt, B.; Lutz, B.; Marsicano, G.; Kokaia, Z.; Guzman, M.; Galve-Roperh, I. The endocannabinoid system drives neural progenitor proliferation. Faseb J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2005, 19, 1704–1706. [Google Scholar] [CrossRef]
- Aguado, T.; Palazuelos, J.; Monory, K.; Stella, N.; Cravatt, B.; Lutz, B.; Marsicano, G.; Kokaia, Z.; Guzman, M.; Galve-Roperh, I. The endocannabinoid system promotes astroglial differentiation by acting on neural progenitor cells. J. Neurosci. 2006, 26, 1551–1561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mulder, J.; Aguado, T.; Keimpema, E.; Barabas, K.; Ballester Rosado, C.J.; Nguyen, L.; Monory, K.; Marsicano, G.; Di Marzo, V.; Hurd, Y.L.; et al. Endocannabinoid signaling controls pyramidal cell specification and long-range axon patterning. Proc. Natl. Acad. Sci. USA 2008, 105, 8760–8765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ayoub, A.E.; Oh, S.; Xie, Y.; Leng, J.; Cotney, J.; Dominguez, M.H.; Noonan, J.P.; Rakic, P. Transcriptional programs in transient embryonic zones of the cerebral cortex defined by high-resolution mRNA sequencing. Proc. Natl. Acad. Sci. USA 2011, 108, 14950–14955. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baker, T.; Datta, P.; Rewers-Felkins, K.; Thompson, H.; Kallem, R.R.; Hale, T.W. Transfer of Inhaled Cannabis Into Human Breast Milk. Obstet. Gynecol. 2018, 131, 783–788. [Google Scholar] [CrossRef] [PubMed]
- Metz, T.D.; Stickrath, E.H. Marijuana use in pregnancy and lactation: A review of the evidence. Am. J. Obstet. Gynecol. 2015, 213, 761–778. [Google Scholar] [CrossRef] [PubMed]
- Morozov, Y.M.; Freund, T.F. Post-natal development of type 1 cannabinoid receptor immunoreactivity in the rat hippocampus. Eur. J. Neurosci. 2003, 18, 1213–1222. [Google Scholar] [CrossRef] [PubMed]
- Morozov, Y.M.; Sun, Y.Y.; Kuan, C.Y.; Rakic, P. Alteration of SLP2-like immunolabeling in mitochondria signifies early cellular damage in developing and adult mouse brain. Eur. J. Neurosci. 2016, 43, 245–257. [Google Scholar] [CrossRef] [Green Version]
- Gal, J.S.; Morozov, Y.M.; Ayoub, A.E.; Chatterjee, M.; Rakic, P.; Haydar, T.F. Molecular and morphological heterogeneity of neural precursors in the mouse neocortical proliferative zones. J. Neurosci. 2006, 26, 1045–1056. [Google Scholar] [CrossRef] [Green Version]
- Tabata, H.; Nakajima, K. Multipolar migration: The third mode of radial neuronal migration in the developing cerebral cortex. J. Neurosci. 2003, 23, 9996–10001. [Google Scholar] [CrossRef] [Green Version]
- Nadarajah, B.; Brunstrom, J.E.; Grutzendler, J.; Wong, R.O.; Pearlman, A.L. Two modes of radial migration in early development of the cerebral cortex. Nat. Neurosci. 2001, 4, 143–150. [Google Scholar] [CrossRef]
- Katona, I.; Rancz, E.A.; Acsady, L.; Ledent, C.; Mackie, K.; Hajos, N.; Freund, T.F. Distribution of CB1 cannabinoid receptors in the amygdala and their role in the control of GABAergic transmission. J. Neurosci. 2001, 21, 9506–9518. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, T.; Maroso, M.; Beer, A.; Baddenhausen, S.; Ludewig, S.; Fan, W.; Vennin, C.; Loch, S.; Berninger, B.; Hofmann, C.; et al. Neural stem cell lineage-specific cannabinoid type-1 receptor regulates neurogenesis and plasticity in the adult mouse hippocampus. Cereb. Cortex (N.Y. 1991) 2018, 28, 4454–4471. [Google Scholar] [CrossRef] [PubMed]
- Zimmer, A.; Zimmer, A.M.; Hohmann, A.G.; Herkenham, M.; Bonner, T.I. Increased mortality, hypoactivity, and hypoalgesia in cannabinoid CB1 receptor knockout mice. Proc. Natl. Acad. Sci. USA 1999, 96, 5780–5785. [Google Scholar] [CrossRef] [Green Version]
- Bacci, A.; Huguenard, J.R.; Prince, D.A. Long-lasting self-inhibition of neocortical interneurons mediated by endocannabinoids. Nature 2004, 431, 312–316. [Google Scholar] [CrossRef] [PubMed]
- Marinelli, S.; Pacioni, S.; Cannich, A.; Marsicano, G.; Bacci, A. Self-modulation of neocortical pyramidal neurons by endocannabinoids. Nat. Neurosci. 2009, 12, 1488–1490. [Google Scholar] [CrossRef] [PubMed]
- Komuro, H.; Kumada, T. Ca2+ transients control CNS neuronal migration. Cell Calcium 2005, 37, 387–393. [Google Scholar] [CrossRef]
- Komuro, H.; Rakic, P. Orchestration of neuronal migration by activity of ion channels, neurotransmitter receptors, and intracellular Ca2+ fluctuations. J. Neurobiol. 1998, 37, 110–130. [Google Scholar] [CrossRef]
- Kapitein, L.C.; Hoogenraad, C.C. Building the Neuronal Microtubule Cytoskeleton. Neuron 2015, 87, 492–506. [Google Scholar] [CrossRef] [Green Version]
- Morozov, Y.M.; Ayoub, A.E.; Rakic, P. Translocation of synaptically connected interneurons across the dentate gyrus of the early postnatal rat hippocampus. J. Neurosci. 2006, 26, 5017–5027. [Google Scholar] [CrossRef] [Green Version]
- Morozov, Y.M.; Koch, M.; Rakic, P.; Horvath, T.L. Cannabinoid type 1 receptor-containing axons innervate NPY/AgRP neurons in the mouse arcuate nucleus. Mol. Metab. 2017, 6, 374–381. [Google Scholar] [CrossRef]
- Bodor, A.L.; Katona, I.; Nyíri, G.; Mackie, K.; Ledent, C.; Hájos, N.; Freund, T.F. Endocannabinoid signaling in rat somatosensory cortex: Laminar differences and involvement of specific interneuron types. J. Neurosci. 2005, 25, 6845–6856. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsou, K.; Brown, S.; Sañudo-Peña, M.C.; Mackie, K.; Walker, J.M. Immunohistochemical distribution of cannabinoid CB1 receptors in the rat central nervous system. Neuroscience 1998, 83, 393–411. [Google Scholar] [CrossRef]
- Morozov, Y.M.; Dominguez, M.H.; Varela, L.; Shanabrough, M.; Koch, M.; Horvath, T.L.; Rakic, P. Antibodies to cannabinoid type 1 receptor co-react with stomatin-like protein 2 in mouse brain mitochondria. Eur. J. Neurosci. 2013, 38, 2341–2348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fiala, J.C.; Harris, K.M. Extending unbiased stereology of brain ultrastructure to three-dimensional volumes. J. Am. Med. Inform. Assoc. 2001, 8, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fiala, J.C. Reconstruct: A free editor for serial section microscopy. J. Microsc. 2005, 218, 52–61. [Google Scholar] [CrossRef]











Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morozov, Y.M.; Mackie, K.; Rakic, P. Cannabinoid Type 1 Receptor is Undetectable in Rodent and Primate Cerebral Neural Stem Cells but Participates in Radial Neuronal Migration. Int. J. Mol. Sci. 2020, 21, 8657. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21228657
Morozov YM, Mackie K, Rakic P. Cannabinoid Type 1 Receptor is Undetectable in Rodent and Primate Cerebral Neural Stem Cells but Participates in Radial Neuronal Migration. International Journal of Molecular Sciences. 2020; 21(22):8657. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21228657
Chicago/Turabian StyleMorozov, Yury M., Ken Mackie, and Pasko Rakic. 2020. "Cannabinoid Type 1 Receptor is Undetectable in Rodent and Primate Cerebral Neural Stem Cells but Participates in Radial Neuronal Migration" International Journal of Molecular Sciences 21, no. 22: 8657. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21228657