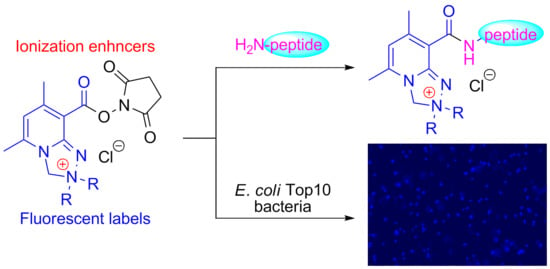
Application of Safirinium N-Hydroxysuccinimide Esters to Derivatization of Peptides for High-Resolution Mass Spectrometry, Tandem Mass Spectrometry, and Fluorescent Labeling of Bacterial Cells
Abstract
:1. Introduction
2. Results and Discussion
2.1. Chemistry
2.2. Mass Spectrometry Analysis
2.2.1. Analysis of Reactive NHS Ester 5a
2.2.2. Analysis of Conjugate 7a, i.e., Peptide Ac-AKF-NH2 Tagged with Diethyl Derivative 5a
2.2.3. Analysis of Conjugate 7b, i.e., Peptide Ac-AKF-NH2 Tagged with Dioctyl Derivative 5b
2.2.4. Analysis of Conjugate 7c, i.e., Peptide Ac-AKF-NH2 Tagged with Pyrrolidine Derivative 5c
2.2.5. Analysis of Conjugate 8, i.e., Tetrapeptide Ac-AAAK Tagged with Diethyl Derivative 5a
2.3. Derivatization of Ubiquitin Hydrolysate with Reagent 5a
2.4. Fluorescence Microscopy
3. Materials and Methods
3.1. General Information
3.2. Chemical Synthesis
3.2.1. Synthesis of 8-Carboxy-5,7-Dimethyl-2,2-Dioctyl-2,3-Dihydro-[1,2,4]Triazolo[4,3-a]Pyridin-2-ium Chloride (3b)
3.2.2. Synthesis of Reactive Safirinium P (5b,c) and Q (6b,c) Probes
3.2.3. Synthesis of Peptide Conjugates 7a-c
3.2.4. Synthesis of the Derivatized Peptide 8
3.3. Tagging the Ubiquitin Hydrolysate with 5a
3.4. Fluorescence Microscopy of Bacteria
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| CID | collision-induced dissociation |
| DCM | dichloromethane |
| EIC | extracted ion chromatogram |
| ESI | electrospray ionization |
| FT-ICR | Fourier-Transform Ion-Cyclotrone-Resonance |
| DIC | N,N-diisopropylcarbodiimide |
| DMF | dimethylformamide |
| DHX | deuterium-hydrogen-exchange |
| GFP | green fluorescence protein |
| HDX | hydrogen-deuterium exchange |
| HPLC | high performance liquid chromatography |
| ITs | ionization tags |
| LB | Luria broth |
| LC | liquid chromatography |
| MRM | multiple reaction monitoring |
| MS | mass spectrometry |
| MS/MS | tandem mass spectrometry |
| NHS | N-hydroxysuccinimide |
| NMR | nuclear magnetic resonance |
| OD | optical density |
| Oct | octyl |
| PAGE | polyacrylamide gel electrophoresis |
| PBS | phosphate-buffered saline |
| QAS | quaternary ammonium salts |
| RP | reverse-phase |
| TEA | triethylamine |
| TEAB | triethylammonium bicarbonate |
| TFA | trifuoroacetic acid |
| TIC | total ion chromatogram |
| TIS | triisopropylsilane |
| TRITC | tetramethylrhodamine-isothiocyanate |
References
- Guzmán-Flores, J.M.; Flores-Pérez, E.C.; Hernández-Ortiz, M.; Vargas-Ortiz, K.; Ramírez-Emiliano, J.; Encarnación-Guevara, S.; Pérez-Vázquez, V. Protein Expression Profile of Twenty-Week-Old Diabetic db/db and Non-Diabetic Mice Livers: A Proteomic and Bioinformatic Analysis. Biomolecules 2018, 8, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagarajan, A.; Zhou, M.; Nguyen, A.Y.; Liberton, M.; Kedia, K.; Shi, T.; Piehowski, P.; Shukla, A.; Fillmore, T.L.; Nicora, C.; et al. Proteomic Insights into Phycobilisome Degradation, A Selective and Tightly Controlled Process in The Fast-Growing Cyanobacterium Synechococcus elongatus UTEX 2973. Biomolecules 2019, 9, 374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, A.-L.; Liu, Y.-N.; Liu, R.; Ren, A.; Ma, H.-Y.; Shu, L.-B.; Shi, L.; Zhu, J.; Zhao, M.-W. Integrated Proteomics and Metabolomics Analysis Provides Insights into Ganoderic Acid Biosynthesis in Response to Methyl Jasmonate in Ganoderma Lucidum. Int. J. Mol. Sci. 2019, 20, 6116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- L’Hocine, L.; Pitre, M.; Achouri, A. Detection and Identification of Allergens from Canadian Mustard Varieties of Sinapis alba and Brassica juncea. Biomolecules 2019, 9, 489. [Google Scholar] [CrossRef] [Green Version]
- Soboleva, A.; Modzel, M.; Didio, A.; Płóciennik, H.; Kijewska, M.; Grischina, T.; Karonova, T.; Bilova, T.; Stefanov, V.; Stefanowicz, P.; et al. Quantification of prospective type 2 diabetes mellitus biomarkers by stable isotope dilution with bi-labeled standard glycated peptides. Anal. Methods 2017, 9, 409–418. [Google Scholar] [CrossRef]
- Verrastro, I.; Pasha, S.; Jensen, K.T.; Pitt, A.R.; Spickett, C.M. Mass Spectrometry-Based Methods for Identifying Oxidized Proteins in Disease: Advances and Challenges. Biomolecules 2015, 5, 378–411. [Google Scholar] [CrossRef] [Green Version]
- Jamnongkan, W.; Lebrilla, C.B.; Barboza, M.; Techasen, A.; Loilome, W.; Sithithaworn, P.; Khuntikeo, N.; Pairojkul, C.; Chamadol, N.; Thanan, R.; et al. Discovery of Serotransferrin Glycoforms: Novel Markers for Diagnosis of Liver Periductal Fibrosis and Prediction of Cholangiocarcinoma. Biomolecules 2019, 9, 538. [Google Scholar] [CrossRef] [Green Version]
- Cebo, M.; Kielmas, M.; Adamczyk, J.; Cebrat, M.; Szewczuk, Z.; Stefanowicz, P. Hydrogen–deuterium exchange in imidazole as a tool for studying histidine phosphorylation. Anal. Bioanal. Chem. 2014, 406, 8013–8020. [Google Scholar] [CrossRef] [Green Version]
- Lu, Z.-S.; Chen, Q.-S.; Zheng, Q.-X.; Shen, J.-J.; Luo, Z.-P.; Fan, K.; Xu, S.-H.; Shen, Q.; Liu, P.-P. Proteomic and Phosphoproteomic Analysis in Tobacco Mosaic Virus-Infected Tobacco (Nicotiana tabacum). Biomolecules 2019, 9, 39. [Google Scholar] [CrossRef] [Green Version]
- Kozuka-Hata, H.; Kitamura, A.; Hiroki, T.; Aizawa, A.; Tsumoto, K.; Inoue, J.-I.; Oyama, M. System-Wide Analysis of Protein Acetylation and Ubiquitination Reveals a Diversified Regulation in Human Cancer Cells. Biomolecules 2020, 10, 411. [Google Scholar] [CrossRef] [Green Version]
- Purushothaman, K.; Das, P.P.; Presslauer, C.; Lim, T.K.; Johansen, S.D.; Lin, Q.; Babiak, I. Proteomics Analysis of Early Developmental Stages of Zebrafish Embryos. Int. J. Mol. Sci. 2019, 20, 6359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kęska, P.; Wójciak, K.M.; Stadnik, J. Effect of Marination Time on the Antioxidant Properties of Peptides Extracted from Organic Dry-Fermented Beef. Biomolecules 2019, 9, 614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Proaño-Bolaños, C.; Blasco-Zúñiga, A.; Almeida, J.R.; Wang, L.; Llumiquinga, M.A.; Rivera, M.; Zhou, M.; Chen, T.; Shaw, C. Unravelling the Skin Secretion Peptides of the Gliding Leaf Frog, Agalychnis spurrelli (Hylidae). Biomolecules 2019, 9, 667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dasgupta, S.; Fishman, M.A.; Castro, L.M.; Tashima, A.K.; Ferro, E.S.; Fricker, L.D. Effect of Protein Denaturation and Enzyme Inhibitors on Proteasomal-Mediated Production of Peptides in Human Embryonic Kidney Cells. Biomolecules 2019, 9, 207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Setner, B.; Stefanowicz, P.; Szewczuk, Z. Quaternary ammonium isobaric tag for a relative and absolute quantification of peptides. J. Mass Spectrom. 2018, 53, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Bąchor, R.; Waliczek, M.; Stefanowicz, P.; Szewczuk, Z. Trends in the Design of New Isobaric Labeling Reagents for Quantitative Proteomics. Molecules 2019, 24, 701. [Google Scholar] [CrossRef] [Green Version]
- Waliczek, M.; Bąchor, R.; Kijewska, M.; Gąszczyk, D.; Panek-Laszczyńska, K.; Konieczny, A.; Dąbrowska, K.; Witkiewicz, W.; Marek-Bukowiec, K.; Tracz, J.; et al. Isobaric duplex based on a combination of 16O/18O enzymatic exchange and labeling with pyrylium salts. Anal. Chim. Acta 2019, 1048, 96–104. [Google Scholar] [CrossRef]
- Bąchor, R.; Dębowski, D.; Łęgowska, A.; Stefanowicz, P.; Rolka, K.; Szewczuk, Z. Convenient preparation of deuterium-labeled analogs of peptides containing N-substituted glycines for a stable isotope dilution LC-MS quantitative analysis. J. Pept. Sci. 2015, 21, 819–825. [Google Scholar] [CrossRef]
- Aoki, M.M.; Kisiala, A.B.; Li, S.; Stock, N.L.; Brunetti, C.R.; Huber, R.J.; Emery, R.J.N. Cytokinin Detection during the Dictyostelium discoideum Life Cycle: Profiles Are Dynamic and Affect Cell Growth and Spore Germination. Biomolecules 2019, 9, 702. [Google Scholar] [CrossRef] [Green Version]
- Modzel, M.; Płóciennik, H.; Kielmas, M.; Szewczuk, Z.; Stefanowicz, P. A synthesis of new, bi-labeled peptides for quantitative proteomics. J. Proteom. 2015, 115, 1–7. [Google Scholar] [CrossRef]
- Bąchor, R.; Kluczyk, A.; Stefanowicz, P.; Szewczuk, Z. Preparation of novel deuterated cyclosporin A standards for quantitative LC-MS analysis. J. Mass Spectrom. 2017, 52, 817–822. [Google Scholar] [CrossRef] [PubMed]
- Slavata, L.; Chmelík, J.; Kavan, D.; Filandrová, R.; Fiala, J.; Rosůlek, M.; Mrázek, H.; Kukačka, Z.; Vališ, K.; Man, P.; et al. MS-Based Approaches Enable the Structural Characterization of Transcription Factor/DNA Response Element Complex. Biomolecules 2019, 9, 535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bąchor, R.; Cydzik, M.; Rudowska, M.; Kluczyk, A.; Stefanowicz, P.; Szewczuk, Z. Sensitive electrospray mass spectrometry analysis of one-bead-one-compound peptide libraries labeled by quaternary ammonium salts. Mol. Divers. 2012, 16, 613–618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bąchor, R.; Mielczarek, P.; Rudowska, M.; Silberring, J.; Szewczuk, Z. Sensitive detection of charge derivatized peptides at the attomole level using nano-LC-ESI–MRM analysis. Int. J. Mass Spectrom. 2014, 362, 32–38. [Google Scholar] [CrossRef]
- Kijewska, M.; Kuc, A.; Kluczyk, A.; Waliczek, M.; Man-Kupisinska, A.; Lukasiewicz, J.; Stefanowicz, P.; Szewczuk, Z. Selective Detection of Carbohydrates and Their Peptide Conjugates by ESI-MS Using Synthetic Quaternary Ammonium Salt Derivatives of Phenylboronic Acids. J. Am. Soc. Mass Spectrom. 2014, 25, 966–976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cydzik, M.; Rudowska, M.; Stefanowicz, P.; Szewczuk, Z. Derivatization of peptides as quaternary ammonium salts for sensitive detection by ESI-MS. J. Pept. Sci. 2011, 17, 445–453. [Google Scholar] [CrossRef] [PubMed]
- Setner, B.; Wierzbicka, M.; Jerzykiewicz, L.; Lisowski, M.; Szewczuk, Z. The unexpected racemization and hydrogen–deuterium exchange of the hydrogen at the α-carbon of proline analogs containing the 5-azoniaspiro[4.4]nonyl-group. Org. Biomol. Chem. 2018, 16, 825–831. [Google Scholar] [CrossRef]
- Setner, B.; Rudowska, M.; Klem, E.; Cebrat, M.; Szewczuk, Z. Peptides derivatized with bicyclic quaternary ammonium ionization tags. Sequencing via tandem mass spectrometry. J. Mass Spectrom. 2014, 49, 995–1001. [Google Scholar] [CrossRef]
- Setner, B.; Rudowska, M.; Kluczyk, A.; Stefanowicz, P.; Szewczuk, Z. The 5-azoniaspiro[4.4]nonyl group for improved MS peptide analysis: A novel non-fragmenting ionization tag for mass spectrometric sensitive sequencing of peptides. Anal. Chim. Acta 2017, 986, 71–81. [Google Scholar] [CrossRef]
- Setner, B.; Szewczuk, Z. New ionization tags based on the structure of the 5-azoniaspiro[4.4]nonyl tag for a sensitive peptide sequencing by mass spectrometry. Anal Bioanal Chem. 2014, 410, 1311–1321. [Google Scholar] [CrossRef] [Green Version]
- Fink, J.; Pathak, H.; Smith, J.; Achat-Mendes, C.; Haining, R.L. Development of a Competition-Binding Assay to Determine Binding Affinity of Molecules to Neuromelanin via Fluorescence Spectroscopy. Biomolecules 2019, 9, 175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodrigues, A.C.B.; Mariz, I.d.F.A.; Maçoas, E.M.S.; Tonelli, R.R.; Martinho, J.M.G.; Quina, F.H.; Bastos, E.L. Bioinspired water-soluble two-photon fluorophores. Dyes Pigment. 2018, 150, 105–111. [Google Scholar] [CrossRef]
- Bodio, E.; Goze, C. Investigation of B-F substitution on BODIPY and aza-BODIPY dyes: Development of B-O and B-C BODIPYs. Dyes Pigment. 2019, 160, 700–710. [Google Scholar] [CrossRef]
- Bernhard, Y.; Richard, P.; Decreau, R.A. Addressing subphthalocyanines and subnaphthalocyanines features relevant to fluorescence imaging. Tetrahedron 2018, 74, 1047–1052. [Google Scholar] [CrossRef]
- Rezende, L.C.D.; Melo, S.M.G.; Boodts, S.; Verbelen, B.; Emery, F.S.; Dehaen, W. Thiocyanation of 3-substituted and 3,5-disubstituted BODIPYs and its application for the synthesis of new fluorescent sensors. Dyes Pigment. 2018, 154, 155–163. [Google Scholar] [CrossRef]
- Pidluzhna, A.; Ivaniuk, K.; Stakhira, P.; Hotra, Z.; Chapran, M.; Ulanski, J.; Tynkevych, O.; Khalavka, Y.; Baryshnikov, G.V.; Minaev, B.F.; et al. Multi-channel electroluminescence of CdTe/CdS core-shell quantum dots implemented into a QLED device. Dyes Pigment. 2019, 162, 647–653. [Google Scholar] [CrossRef]
- Oliveira, E.; Santos, H.M.; Jorge, S.; Rodríguez-González, B.; Novio, F.; Lorenzo, J.; Ruiz-Molina, D.; Luis Capelo, J.; Lodeiro, C. Sustainable synthesis of luminescent CdTe quantum dots coated with modified silica mesoporous nanoparticles: Towards new protein scavengers and smart drug delivery carriers. Dyes Pigment. 2019, 161, 360–369. [Google Scholar] [CrossRef] [Green Version]
- Zahid, M.; Feldman, K.S.; Garcia-Borrero, G.; Feinstein, T.N.; Pogodzinski, N.; Xu, X.; Yurko, R.; Czachowski, M.; Wu, Y.L.; Mason, N.S.; et al. Cardiac Targeting Peptide, a Novel Cardiac Vector: Studies in Bio-Distribution, Imaging Application, and Mechanism of Transduction. Biomolecules 2018, 8, 147. [Google Scholar] [CrossRef] [Green Version]
- Kiyama, M.; Iwano, S.; Otsuka, S.; Lu, S.W.; Obata, R.; Miyawaki, A.; Hirano, T.; Maki, S.A. Quantum yield improvement of red-light-emitting firefly luciferin analogues for in vivo bioluminescence imaging. Tetrahedron 2018, 74, 652–660. [Google Scholar] [CrossRef]
- Hanif, M.; Rafiq, M.; Yousuf, M.; Kotwica-Mojzych, K.; Saleem, M.; Mojzych, M. Organic small molecular receptors as fluorimetric/bioimaging probe for extracellular/intracellular zinc sensation. Bioorg. Chem. 2020, 94, 103398. [Google Scholar] [CrossRef]
- Váradi, J.; Hermenean, A.; Gesztelyi, R.; Jeney, V.; Balogh, E.; Majoros, L.; Malanga, M.; Fenyvesi, É.; Szente, L.; Bácskay, I.; et al. Pharmacokinetic Properties of Fluorescently Labelled Hydroxypropyl-Beta-Cyclodextrin. Biomolecules 2019, 9, 509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saczewski, J.; Hinc, K.; Obuchowski, M.; Gdaniec, M. The tandem Mannich–electrophilic amination reaction: A versatile platform for fluorescent probing and labelling. Chem. Eur. J. 2013, 19, 11531–11535. [Google Scholar] [CrossRef] [PubMed]
- Fedorowicz, J.; Sączewski, J.; Drażba, Z.; Wiśniewska, P.; Gdaniec, M.; Wicher, B.; Suwiński, G.; Jalińska, A. Synthesis and fluorescence of dihydro-[1,2,4]triazolo[4,3-a]pyridin-2-iumcarboxylates: An experimental and TD-DFT comparative study. Dyes Pigment. 2019, 161, 347–359. [Google Scholar] [CrossRef]
- Fedorowicz, J.; Sączewski, J.; Konopacka, A.; Waleron, K.; Lejnowski, D.; Ciura, K.; Tomasic, T.; Skok, Z.; Savijoki, K.; Morawska, M.; et al. Synthesis and biological evaluation of hybrid quinolone-based quaternary ammonium antibacterial agents. Eur. J. Med. Chem. 2019, 179, 576–590. [Google Scholar] [CrossRef] [PubMed]
- Ciura, K.; Fedorowicz, J.; Andrić, F.; Greber, K.E.; Gurgielewicz, A.; Sawicki, W.; Sączewski, J. Lipophilicity Determination of Quaternary (Fluoro) Quinolones by Chromatographic and Theoretical Approaches. Int. J. Mol. Sci. 2019, 20, 5288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ciura, K.; Fedorowicz, J.; Kapica, H.; Adamkowska, A.; Sawicki, W.; Sączewski, J. Affinity of Fluoroquinolone–Safirinium Dye Hybrids to Phospholipids Estimated by IAM-HPLC. Processes 2020, 8, 1148. [Google Scholar] [CrossRef]
- Kraft, O.; Kozubek, M.; Hoenke, S.; Serbian, I.; Major, D.; Csuk, R. Cytotoxic triterpenoid-safirinium conjugates target the endoplasmic reticulum. Eur. J. Med. Chem. 2021, 209, 112920. [Google Scholar] [CrossRef]
- Fedorowicz, J.; Cebrat, M.; Wierzbicka, M.; Wiśniewska, P.; Jalińska, A.; Dziomba, S.; Gdaniec, M.; Jaremko, M.; Jaremko, Ł.; Chandra, K.; et al. Synthesis and evaluation of dihydro-[1,2,4]triazolo[4,3-a]pyridin-2-ium carboxylates as fixed charge fluorescent derivatization reagents for MEKC and MS proteomic analyses. J. Mol. Struct. 2020, 1217, 128426. [Google Scholar] [CrossRef]
- Cydzik, M.; Rudowska, M.; Stefanowicz, P.; Szewczuk, Z. The Competition of Charge Remote and Charge Directed Fragmentation Mechanisms in Quaternary Ammonium Salt Derivatized Peptides—An Isotopic Exchange Study. J. Am. Soc. Mass Spectrom. 2011, 22, 2103–2107. [Google Scholar] [CrossRef] [Green Version]
- Gross, M.L. Charge-remote fragmentations: Method, mechanism and applications. Int. J. Mass Spectrom. Ion Process. 1992, 118–119, 137–165. [Google Scholar] [CrossRef]
- Demarque, D.P.; Crotti, A.E.M.; Vessecchi, R.; Lopes, J.L.C.; Lopes, N.P. Fragmentation reactions using electrospray ionization mass spectrometry: An important tool for the structural elucidation and characterization of synthetic and natural products. Nat. Prod. Rep. 2016, 33, 432–455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Setner, B.; Rudowska, M.; Wojewska, D.; Kluczyk, A.; Stefanowicz, P.; Szewczuk, Z. Peptides labeled by 5-azoniaspiro[4.4]nonyl group for sensitive sequencing by electrospray tandem mass spectrometry. J. Pept. Sci. 2014, 20, S64–S65. [Google Scholar] [CrossRef]
- Syka, J.E.P.; Coon, J.J.; Schroeder, M.J.; Shabanowitz, J.; Hunt, D.F. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc. Natl. Acad. Sci. USA 2004, 101, 9528–9533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaremko, Ł.; Jaremko, M.; Pasikowski, P.; Cebrat, M.; Stefanowicz, P.; Lisowski, M.; Artym, J.; Zimecki, M.; Zhukov, I.; Szewczuk, Z. The immunosuppressive activity and solution structures of ubiquitin fragments. Biopolymers 2009, 91, 423–431. [Google Scholar] [CrossRef]
- Sączewski, J.; Fedorowicz, J.; Korcz, M.; Sączewski, F.; Wicher, B.; Gdaniec, M.; Konopacka, A. Experimental and theoretical studies on the tautomerism and reactivity of isoxazolo[3,4-b]quinolin-3(1H)-ones. Tetrahedron 2015, 71, 8975–8984. [Google Scholar] [CrossRef]



















| No | Peptide Sequence | [M+H]+ | [M+Saf]1+ | [M+2H]2+ | [M+H+Saf]2+ | [M+2Saf]2+ | [M+3H]3+ | [M+2H+Saf]3+ | [M+H+2Saf]3+ | [M+3Saf]3+ | [M+4H]4+ | [M+4Saf]4+ |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | NVKAKIQDKEGIPPDQQRL | - | - | - | - | 726.4061 | - | - | - | 545.0564 | - | |
| 2 | NVKAKIQDKEGIPPDQQ | - | 954.5129 | - | - | - | - | - | - | - | - | |
| 3 | NVKAKIQDKEGIPPDQ | - | 890.4837 | - | - | - | - | - | - | - | 676.8827 | |
| 4 | AGKQLEDGRTLSD | - | 695.3521 | - | 926.4893 | - | 540.9496 | 617.9953 | - | - | ||
| 5 | VKTLTGKTITL | - | 587.8740 | - | - | - | - | - | 623.3889 | - | - | |
| 6 | YNIQKESTL | 1095.5681 | - | 548.2877 | - | 779.4249 | - | - | - | 596.9981 | - | - |
| 7 | YNIQKEST | 982.4840 | - | - | - | 722.8829 | - | - | - | - | - | - |
| 8 | NIQKESTL | - | - | - | 582.3246 | 697.8932 | - | - | - | - | - | - |
| 9 | VKTLTGKT | 847.5248 | - | 424.2661 | - | - | - | - | - | 514.3170 | - | - |
| 10 | AGKQLEDG | 817.4051 | - | - | - | 640.3434 | - | - | - | - | - | - |
| 11 | AGKQLED | - | - | - | - | 611.8326 | - | - | - | - | - | - |
| 12 | QRLIF | 676.4141 | - | - | - | - | - | - | - | - | - | - |
| 13 | EVEPSD | 675.2832 | 906.4204 | - | - | - | - | - | - | - | - | - |
| 14 | VKTLTG | 618.3821 | - | - | - | 540.8319 | - | - | - | - | - | - |
| 15 | KESTL | 577.3192 | - | - | - | 520.3004 | - | - | - | - | - | - |
| 16 | RLRGG | 558.3471 | - | - | - | - | - | - | - | - | - | - |
| 17 | MQIF | 538.2694 | 769.4066 | - | - | - | - | - | - | - | - | - |
| 18 | HLVL | 481.3133 | 712.4505 | - | - | 472.2975 | - | - | - | - | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fedorowicz, J.; Wierzbicka, M.; Cebrat, M.; Wiśniewska, P.; Piątek, R.; Zalewska-Piątek, B.; Szewczuk, Z.; Sączewski, J. Application of Safirinium N-Hydroxysuccinimide Esters to Derivatization of Peptides for High-Resolution Mass Spectrometry, Tandem Mass Spectrometry, and Fluorescent Labeling of Bacterial Cells. Int. J. Mol. Sci. 2020, 21, 9643. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21249643
Fedorowicz J, Wierzbicka M, Cebrat M, Wiśniewska P, Piątek R, Zalewska-Piątek B, Szewczuk Z, Sączewski J. Application of Safirinium N-Hydroxysuccinimide Esters to Derivatization of Peptides for High-Resolution Mass Spectrometry, Tandem Mass Spectrometry, and Fluorescent Labeling of Bacterial Cells. International Journal of Molecular Sciences. 2020; 21(24):9643. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21249643
Chicago/Turabian StyleFedorowicz, Joanna, Magdalena Wierzbicka, Marek Cebrat, Paulina Wiśniewska, Rafał Piątek, Beata Zalewska-Piątek, Zbigniew Szewczuk, and Jarosław Sączewski. 2020. "Application of Safirinium N-Hydroxysuccinimide Esters to Derivatization of Peptides for High-Resolution Mass Spectrometry, Tandem Mass Spectrometry, and Fluorescent Labeling of Bacterial Cells" International Journal of Molecular Sciences 21, no. 24: 9643. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21249643