3.1. The Global Characteristics
To characterize the statistical and dynamical behavior of the dendrimer as a whole we calculated its size and shape as well as the time correlation functions of the size fluctuation and the rotation as a whole. The mean-squared radius of gyration
is one of the parameters for estimating the characteristic size of the dendrimer. We can obtain
using static light scattering, small angle neutron scattering and small angle X-ray scattering. In simulation
can be calculated as
where
are the molecular masses of the dendrimer and its
i-th atom, correspondingly, and
is the distance from the
i-th atom to the center of mass of the dendrimer. In accordance with the physical meaning of
(Equation (
1)), it provides information about the distribution of the mass of a dendrimer around its center of mass (the larger
, the lower the rotation time of the dendrimer as a whole).
Equation (
1) was used to calculate the instant size of the dendrimer in each time moment (saved every 100 ps in corresponding frames of 1000 ns trajectory file). The time dependence of the radius of gyration shows the pulsation of the dendrimer size (see
Figure 2a). It can be seen from that the size fluctuates between 1.7 nm and 2.3 nm. These fluctuations practically do not depend on temperature (not shown) and are similar to those of the Lys-2Lys dendrimer [
61].
The fluctuations in the radius of gyration indicate that the dendrimer is not a rigid spherical object with a constant radius, but is a molecule with a radius that pulsate in a rather wide range. This pulsating process can be described using the time autocorrelation function that characterizes the correlation of dendrimer sizes
The comparison of the functions
for Lys-2Arg (solid black curve) and for Lys-2Lys (dashed black curve) is shown in
Figure 2b. It is easy to see that this function for Lys-2Arg decreases with time slower than for the Lys-2Lys dendrimer. We will discuss this question in more detail later.
The rotation mobility of the dendrimer can be estimated using the first-order orientational autocorrelation function (ACF)
and the second-order orientational ACF
of the core-to-end vector (the vectors
connect the first branching point in the core of the dendrimer and the C atoms of its terminal NH
groups). To obtain better statistical results we calculated the orientational ACFs (Equations (
3) and (
4)) for each of sixteen
vectors (to each terminal group). The averaged time dependencies of these ACFs for Lys-2Arg and Lys-2Lys are shown in
Figure 2b. We can see the similar behavior of these curves.
The rotation time of the dendrimer as a whole can be estimated as time
where the function
decays in
e times. The rotation times
obtained for the dendrimer as a whole are presented in
Table 2. For both dendrimers, the rotation time decreases with increasing temperature. The difference in the rotation times of these two dendrimers does not exceed 10 percent, which is close to the calculation error of this value. This means that the global rotational motion of the dendrimer as a whole is very similar for both dendrimers.
For discussion of the rotation of the dendrimer as a whole, it is important to know the shape of the rotating object. The shape of the object is also important in practical applications. For example, it is well known that the penetration of rod-shaped molecules and spherical molecules through cell membranes is different [
78]. Many simulation works have shown that the dendrimers of the small generations are asymmetric, but become more spherical as the number of generations increases [
79,
80,
81]. The dendrimer shape can be evaluated using the asphericity parameter
according to the following formula [
80,
82,
83]
where values
,
,
are the eigenvalues of the gyration tensor which are equivalent to ellipsoid axes of a prolate or oblate molecule. In the case of a very prolate molecule, for example, for a rod-like molecule, one axis dominates over the others, and the asphericity in this limiting case tends to unity, while for a spherical molecule
is close to zero.
Table 3 shows the value of
for the Lys-2Lys and Lys-2Arg dendrimers obtained from MD simulation at
K.
We obtained that the value of
for both dendrimers is equal to 0.02, which is very close to 0. It means that we can consider our dendrimers as spherical molecules. We have used the radius of gyration
to characterize the spherical molecule. The temperature dependence of
is presented in
Figure 3a. Also, this figure shows the standard deviations from the equilibrium size in the form of errorbars. It can be seen that the dendrimer size
is practically independent of temperature.
The hydrodynamic radius
is another characteristic of the molecule size that can be measured experimentally. The hydrodynamic radius
is usually estimated as the Stokes radius, i.e., it can be calculated using the coefficient of translational diffusion of the center of mass of the dendrimer. Here, we estimated the hydrodynamic radius of the dendrimer from MD simulation using the Kirkwood approximation [
84,
85]:
where
is a distance between two atoms
i and
j. The proposed formula can be utilized in several ways. In some works on MD simulation of peptides and proteins only C
carbon atoms of the main peptide chain are used. Other methods take into account all heavy atoms in the backbone of a peptide or peptide dendrimer. In addition, all heavy dendrimer atoms and ions or even all heavy dendrimer atoms, ions and oxygen in water molecules that are close to the dendrimer atoms can be considered. We used all of these approaches and found the characteristic ratio
for the Lys-2Arg dendrimer as a function of temperature (
Figure 3b). In
Figure 3b we also indicate two theoretical limits—for a penetrable gaussian coil and for an unpenetrable rigid sphere [
86]. In all cases the obtained MD data are between two limiting values for these theoretical models. An increase in the number of types of heavy atoms that are taken into account when calculating the
using Equation (
6) leads to a better description of the MD data by the unpenetrable sphere model (upper dashed line).
In addition to and , it is important to estimate the position of the outer boundary of the dendrimer in the solvent (i.e., the radial distance between the center of the dendrimer and its spherical surface). This parameter provides a good estimation of the size of the dendrimer as a nanocontainer.
The position of the outer boundary of the sphere can be calculated theoretically as
or through the estimation of the position of the terminal groups
. We can calculate
as the mean square radial distance from the center of the dendrimer to the N atoms at the terminal NH
groups:
Moreover, in simulation, we can use the effective radius
of the dendrimer as a charged macroion.
is estimated from the position of the slip plane and will be determined below from the electrostatic properties of the dendrimer. All of these characteristics (
,
,
) are given in
Table 3. It is interesting to note that the value of
is close to the theoretical value of the radius of the rigid sphere equal to
.
3.3. The Mobility Characteristics Measured in NMR
In this subsection, we calculate the orientational mobility of the specific CH
groups chemically connected to N atoms (marked by dashed circles in
Figure 8). In NMR experiment the signals of CH2-N groups can be measured separately. These NMR active CH
groups belong to three different types of aminoacid residues of Lys-2Arg dendrimer(inner Lys (branching points) (
Figure 8a), side Arg (spacers) (
Figure 8b) and terminal Lys (
Figure 8c). The NMR data for these types of CH
groups in the Lys-2Arg dendrimer were obtained previously [
53].
To obtain the orientational mobility of the H-H vector in the CH
groups (marked by dashed circles in
Figure 8) from MD simulation, we calculated the second-order autocorrelation function ACF
(Equation (
4)) for the H-H vectors in these groups by the same way as we did it for the Lys-2Lys dendrimer in [
61]. The time dependencies of ACFs
for the H-H vectors in three different CH
groups located at inner Lys (
Figure 8a), side Arg (
Figure 8b) and terminal Lys segments (
Figure 8c) are shown in
Figure 9a–c.
Figure 9d demonstrates ACFs
for the core-to-end vectors. The time dependencies were calculated at different temperatures in the range from 280 K to 340 K.
It easy to see that
for all vectors decrease faster with increasing temperature. The
for the H-H vectors of the inner CH
groups decrease very slowly in comparison to the same curves for the terminal CH
groups. However, in the Lys-2Arg dendrimer the
for the H-H vectors of the side CH
groups is close to the
for the inner CH
groups. This fact is in contrast to the results for the Lys-2Lys dendrimer [
52,
61] where
functions for the side and terminal CH
groups are practically the same. The
functions for the core-to-end vectors (
Figure 9d) decay at least ten times slower than ones for the inner CH
groups (
Figure 9a). In addition to ACF
, for the core-to-end vector, we calculated ACF
and
. In several papers on the simulation of polymers and dendrimers [
43,
61,
97] it was shown that there is a simple relationship between the 1st order ACF
and the 2nd order ACF
:
As we can see from
Figure 9d the time dependencies of
and
almost coincide at all temperatures. Thus the relationship (
11) is valid for the core-to-end vector of the Lys-2Arg dendrimer as in other lysine based dendrimers studied earlier.
To compare the mobility characteristics
obtained from MD simulation and NMR experiments we calculated the reduced spin-lattice relaxation rate
in the susceptibility representation. This characteristic is a function of the variable
(the angular frequency of NMR spectrometer) and is calculated using a linear combination of the spectral densities
and
[
98,
99]:
The spectral density is calculated by the cosine Fourier transform
The frequency dependencies of
at different temperatures are presented in
Figure 10. The vertical dashed line in these plots corresponds to the frequency (
MHz) of the spectrometer [
52,
53]. We can see from these plots that frequency dependencies of
change with temperature. Similarly to the time dependencies, these dependencies for the side groups of 2Arg spacers (
Figure 10b) are very similar to
dependencies for the inner groups (
Figure 10a) and differ significantly from those for the terminal ones (
Figure 10c). At the same time, for the Lys-2Lys dendrimer studied earlier [
52,
61], the
dependencies for the side groups of 2Lys spacers were close to the
dependencies for the terminal groups. This large difference in the
behavior of these dendrimers obtained from our MD simulations is very strange. However, this is in good agreement with the NMR experimental results [
52,
53]. We will discuss this difference in detail later in this paper.
In the NMR experiment, the temperature dependence of
is measured at the given frequency of the spectrometer. Therefore, in order to obtain a similar dependence from the simulation, we calculate the position of
at the frequency of the spectrometer at each temperature. We converted the dimensionless of the reduced spin-lattice relaxation rate
to the susceptibility representation of the
(measured in the NMR experiment) using the following relationship:
where
is the constant determined by the quantum chemistry parameters and does not depend on frequency or temperature. The theoretical value of
for CH
groups
is equal to
s
. At the same time,
is often used as a fitting parameter (see for example [
43,
61]). For calculation of the temperature dependence of
we used the theoretical value
for all CH
groups except the terminal CH
groups of Lys-2Arg (for which the value
s
was used).
The temperature dependencies of
for the CH
groups in the Lys-2Arg and Lys-2Lys dendrimers are plotted in
Figure 11. The new simulation results and the experimental NMR data for three types of the CH
groups (inner, side and terminal) in the Lys-2Arg dendrimer [
53] are in very good agreement
Figure 11a. The temperature dependencies of
for the side CH
groups in the Lys-2Arg dendrimer differ from those for the terminal CH
groups. At the same time in our previous NMR and simulation papers [
52,
61], for the Lys-2Lys dendrimer the temperature dependencies of
for the side and terminal CH
groups were practically the same. However, in MD simulation we can calculate the mobility of the terminal and side groups of the Lys-2Lys dendrimer separately. We compared the simulation data both for the Lys-2Arg and Lys-2Lys dendrimers in
Figure 11b. We can conclude that the temperature dependency of
of the side CH
groups in 2Lys spacers of the Lys-2Lys dendrimer is close to those of the terminal CH
groups of both dendrimers. However, the mobility of the side CH
groups of 2Arg spacers of the Lys-2Arg dendrimer is close to the mobility of the inner CH
groups of both dendrimers. Thus, the difference between the mobility of the side CH
groups in 2Arg and 2Lys spacers is not only quantitative but also qualitative. To understand the reason of this difference we performed the calculation of the mobility characteristics for all CH
groups in spacers.
3.4. The Mobility Characteristics of All CH Groups in Side Segments of Spacers from MD Simulation
From previous papers on NMR and MD simulation of different lysine dendrimer [
43,
61], it was established that the mobility of the inner CH
groups significantly differs from the mobility of the terminal groups in the same dendrimers. In [
53] and in the present paper we have obtained a similar result for the Lys-2Arg dendrimer both from the NMR experiment and from the simulation. The mobility of the CH
groups of the Lys-2Lys dendrimers was practically the same as the mobility of the terminal groups [
52,
61]. However, in the Lys-2Arg dendrimer both the NMR experiment [
53] and the MD simulation demonstrate that the mobility of the side CH
groups significantly differ from the mobility of the terminal CH
groups and is close to the mobility of the inner CH
groups.
It was assumed [
53] that a possible reason for this difference in the mobility of the side segments in the Lys-2Arg and Lys-2Lys dendrimers [
52,
53] could be the arginine-arginine pairing [
57,
58], which leads to cross-linking of the Lys-2Arg dendrimer branches. The Arg-Arg pairing effect is well known for arginine dimers and short linear arginine homopeptides. However, it is not clear whether it can play a significant role in the case of dendrimers or not. Another possible reason is that the NMR active CH
groups (C
H
in
Figure 8b) in 2Arg spacers of Lys-2Arg and CH
groups in 2Lys spacers (C
H
in
Figure 8c) in Lys-2Lys occupy the different structural positions from the ends of the side segments (the topological distance). In other words, the topological distance from the C
H
group to the end of the side Arg segment (three bonds) is not the same as the distance from the C
H
group to the end of side Lys segment (two bonds).
If the difference between the mobility of the side segments of Lys-2Arg and Lys-2Lys is due to the Arg-Arg pairing or other types of interaction that lead to effective cross-linking of the neighboring branches of Lys-2Arg, then the orientational mobility of the different CH groups of the side segments will be approximately the same. Otherwise, we can observe the dependence of the mobility of the different CH groups in the same side segment (Arg or Lys) on the distance from a particular CH group to the end of this side segment.
To understand the reason for the different mobility of the NMR active CH
groups, from the MD simulation we calculated the mobility of all CH
groups in each Arg (C
H
, C
H
, C
H
,
Figure 8b) and Lys segment (C
H
, C
H
, C
H
, C
H
,
Figure 8d) of spacers.
To characterize the mobility of these additional CH
groups we calculated the 2nd order ACF
. The results of these calculations are presented in
Figure 12a. It can be seen that the time dependencies of
for the CH
groups located at different topological distances from the end of the side segment in the Lys-2Arg (solid lines) and Lys-2Lys (dashed lines) dendrimers differ. It is important to note that the
for the CH
groups of these dendrimers located at the same topological distance (marked by the same color in
Figure 12a and in
Figure 8) practically coincide. The first of these two results means that our suggestion about the possible cross-linking of dendrimer branches due to the arginine-arginine pairing is not valid (otherwise,
for all CH
groups in spacers of the same dendrimer should be approximatly the same).
The second result means that the topological distance is the main parameter which determines the NMR relaxation of the H-H vectors in CH groups of both spacers.
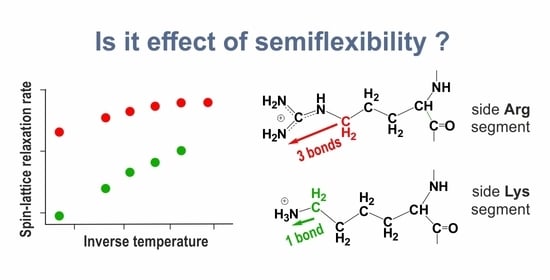
For illustration of these results, we performed a rough evaluation of the relaxation times for each CH
group in 2Arg and 2Lys spacers of the dendrimers under study. In
Table 6, the results of this evaluation are presented as a function of the number of chemical bonds from the C atom of a given CH
group to the N atom of the end NH
(for Arg) or the end NH
group (for Lys) in the side segments of spacers in the Lys-2Arg or Lys-2Lys dendrimers, correspondingly. In the side segments of 2Arg spacers, there are the CH
groups located at the topological distances equal to the length of 3, 4 and 5 chemical bonds. In the side segments of 2Lys spacers, there are the CH
groups located at the topological distances equal to the length of 1, 2, 3, and 4 chemical bonds. The shortest and longest (the statistical error is about 10%) characteristic times for the terminal and inner CH
groups in each dendrimer are presented in the second and last columns in
Table 6, respectively. It is easy to see that characteristic times arranged in this way are very similar for both dendrimers.
In NMR experiments [
52,
53] we considered the side CH2 groups that remoted from the end of the side segment at the distance of one (for Lys-2Lys) and three (for Lys-2Arg) chemical bonds. Therefore, the reason for the difference in the spin-lattice relaxation rates of these side groups measured by NMR [
52,
53] is the different topological distance.
To compare our simulation results and the experimental NMR data for all CH
groups in side segments of 2Arg and 2Lys spacers of both dendrimers, we converted the calculated time dependencies of
to the frequency dependencies of
(using Equation (
12)). The frequency dependencies of
for these CH
groups are shown in
Figure 12b. We can see that the frequency dependencies of
for the CH
groups with the different structural positions differ within the same dendrimer. However, the frequency dependencies of
for the CH
groups located at the same topological distance from the ends of the side segments in these two dendrimers practically coincide. This result confirms our conclusion, based on the comparison of the time dependencies of
for the NMR active CH
groups in 2Arg and 2Lys spacers, that the difference in the mobility of these groups is due to their different topological positions from the end of the corresponding side segment.
Finally, we calculated the temperature dependencies of the spin-lattice NMR relaxation rate
for all side CH
groups in 2Arg and 2Lys spacers of both dendrimers by analogy with [
52,
53]. These dependencies are presented in
Figure 13a for the Lys-2Arg dendrimer and in
Figure 13b for the Lys-2Lys dendrimer. According to these figures, in general, the temperature dependencies of
looks rather similar for both dendrimers if we compare, for instance, the curves for the side CH
-N groups.
We see that with distance from the end of the side segment, the mobility decreases. This behavior is related to the NMR semiflexibility effect of hyperbranched macromolecules [
100,
101,
102] and is discussed in some detail for lysine dendrimers [
43].