FRCaMP, a Red Fluorescent Genetically Encoded Calcium Indicator Based on Calmodulin from Schizosaccharomyces Pombe Fungus
Abstract
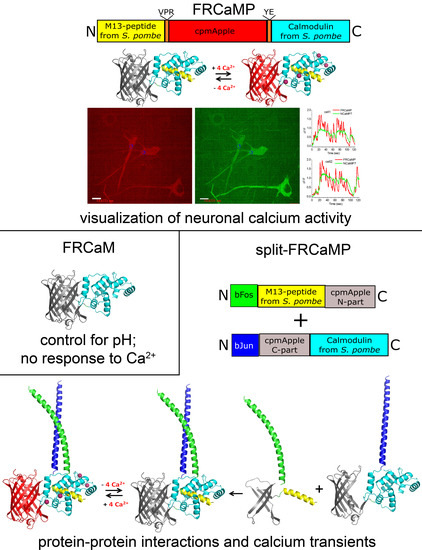
:1. Introduction
2. Results and Discussion
2.1. Developing a Red Calcium Indicator Based on cpmApple and CaM and the M13-Like Peptide from S. pombe Fungus in a Bacterial System
2.2. In Vitro Characterization of the Purified FRCaMP Indicator
2.3. Calcium-Dependent Response of the FRCaMP Calcium Indicator in HeLa Mammalian Cells
2.4. Visualization of Spontaneous Neuronal Activity in the Dissociated Culture Using the FRCaMP Indicator and Confocal Imaging
2.5. Visualization of Induced Neuronal Activity in Dissociated Culture Using the FRCaMP Indicator and Confocal Imaging
2.6. Characterization of Truncated Versions (with a Deleted M13-Like Peptide) of the FRCaMP and GCaMP6s Indicators In Vitro and in HeLa Cells
2.7. Generation and Characterization of the Split-Version of the FRCaMP Indicator in HeLa Cells
3. Materials and Methods
3.1. Mutagenesis and Library Screening
3.2. Protein Purification and Characterization
3.3. Bacterial and Mammalian Plasmids Construction
3.4. Mammalian Live-Cell Imaging
3.5. Imaging in Primary Mouse Neuronal Cultures
3.6. Statistics
3.7. Ethical Approval and Animal Care
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| MDPI | Multidisciplinary Digital Publishing Institute |
| GECI | Genetically encoded calcium indicator |
| FP | Fluorescent protein |
| rAAV | Recombinant adeno-associated virus |
| DIVs | Days in vitro |
| HEPES | 2-[4-(2-hydroxyethyl)piperazin-1-yl]ethanesulfonic acid |
| MOPS | 3-(N-morpholino)propanesulfonic acid |
| QY | Quantum yield |
| AP | Action Potential |
| SD | Standard deviation |
| CNQX | 6-cyano-7-nitroquinoxaline-2,3-dione |
| APV or AP5 | (2R)-amino-5-phosphonovaleric acid |
References
- Piatkevich, K.D.; Murdock, M.H.; Subach, F.V. Advances in Engineering and Application of Optogenetic Indicators for Neuroscience. Appl. Sci. 2019, 9, 562. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez, E.A.; Campbell, R.E.; Lin, J.Y.; Lin, M.Z.; Miyawaki, A.; Palmer, A.E.; Shu, X.; Zhang, J.; Tsien, R.Y. The Growing and Glowing Toolbox of Fluorescent and Photoactive Proteins. Trends Biochem. Sci. 2017, 42, 111–129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barykina, N.V.; Sotskov, V.P.; Gruzdeva, A.M.; Wu, Y.K.; Portugues, R.; Subach, O.M.; Chefanova, E.S.; Plusnin, V.V.; Ivashkina, O.I.; Anokhin, K.V.; et al. FGCaMP7, an Improved Version of Fungi-Based Ratiometric Calcium Indicator for In Vivo Visualization of Neuronal Activity. Int. J. Mol. Sci. 2020, 21, 3012. [Google Scholar] [CrossRef]
- Barykina, N.V.; Subach, O.M.; Piatkevich, K.D.; Jung, E.E.; Malyshev, A.Y.; Smirnov, I.V.; Bogorodskiy, A.O.; Borshchevskiy, V.I.; Varizhuk, A.M.; Pozmogova, G.E.; et al. Green fluorescent genetically encoded calcium indicator based on calmodulin/M13-peptide from fungi. PLoS ONE 2017, 12, e0183757. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Araki, S.; Wu, J.; Teramoto, T.; Chang, Y.F.; Nakano, M.; Abdelfattah, A.S.; Fujiwara, M.; Ishihara, T.; Nagai, T.; et al. An expanded palette of genetically encoded Ca(2) indicators. Science 2011, 333, 1888–1891. [Google Scholar] [CrossRef] [Green Version]
- Shen, Y.; Dana, H.; Abdelfattah, A.S.; Patel, R.; Shea, J.; Molina, R.S.; Rawal, B.; Rancic, V.; Chang, Y.F.; Wu, L.; et al. A genetically encoded Ca(2+) indicator based on circularly permutated sea anemone red fluorescent protein eqFP578. BMC Biol. 2018, 16, 9. [Google Scholar] [CrossRef] [Green Version]
- Dana, H.; Mohar, B.; Sun, Y.; Narayan, S.; Gordus, A.; Hasseman, J.P.; Tsegaye, G.; Holt, G.T.; Hu, A.; Walpita, D.; et al. Sensitive red protein calcium indicators for imaging neural activity. Elife 2016, 5, e12727. [Google Scholar] [CrossRef]
- Barykina, N.V.; Doronin, D.A.; Subach, O.M.; Sotskov, V.P.; Plusnin, V.V.; Ivleva, O.A.; Gruzdeva, A.M.; Kunitsyna, T.A.; Ivashkina, O.I.; Lazutkin, A.A.; et al. NTnC-like genetically encoded calcium indicator with a positive and enhanced response and fast kinetics. Sci. Rep. 2018, 8, 15233. [Google Scholar] [CrossRef] [Green Version]
- Subach, F.V.; Piatkevich, K.D.; Verkhusha, V.V. Directed molecular evolution to design advanced red fluorescent proteins. Nat. Methods 2011, 8, 1019–1026. [Google Scholar] [CrossRef]
- Shaner, N.C.; Lin, M.Z.; McKeown, M.R.; Steinbach, P.A.; Hazelwood, K.L.; Davidson, M.W.; Tsien, R.Y. Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat. Methods 2008, 5, 545–551. [Google Scholar] [CrossRef] [Green Version]
- Akerboom, J.; Carreras Calderon, N.; Tian, L.; Wabnig, S.; Prigge, M.; Tolo, J.; Gordus, A.; Orger, M.B.; Severi, K.E.; Macklin, J.J.; et al. Genetically encoded calcium indicators for multi-color neural activity imaging and combination with optogenetics. Front. Mol. Neurosci. 2013, 6, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shaner, N.C.; Campbell, R.E.; Steinbach, P.A.; Giepmans, B.N.; Palmer, A.E.; Tsien, R.Y. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 2004, 22, 1567–1572. [Google Scholar] [CrossRef] [PubMed]
- Subach, O.M.; Cranfill, P.J.; Davidson, M.W.; Verkhusha, V.V. An enhanced monomeric blue fluorescent protein with the high chemical stability of the chromophore. PLoS ONE 2011, 6, e28674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heim, R.; Cubitt, A.B.; Tsien, R.Y. Improved green fluorescence. Nature 1995, 373, 663–664. [Google Scholar] [CrossRef]
- Kaushik, S.; Cuervo, A.M. The coming of age of chaperone-mediated autophagy. Nat. Rev. Mol. Cell Biol. 2018, 19, 365–381. [Google Scholar] [CrossRef] [PubMed]
- Davis, A.A.; Leyns, C.E.G.; Holtzman, D.M. Intercellular Spread of Protein Aggregates in Neurodegenerative Disease. Annu. Rev. Cell Dev. Biol. 2018, 34, 545–568. [Google Scholar] [CrossRef]
- Wu, J.; Abdelfattah, A.S.; Miraucourt, L.S.; Kutsarova, E.; Ruangkittisakul, A.; Zhou, H.; Ballanyi, K.; Wicks, G.; Drobizhev, M.; Rebane, A.; et al. A long Stokes shift red fluorescent Ca(2+) indicator protein for two-photon and ratiometric imaging. Nat. Commun. 2014, 5, 5262. [Google Scholar] [CrossRef]
- Subach, O.M.; Sotskov, V.P.; Plusnin, V.V.; Gruzdeva, A.M.; Barykina, N.V.; Ivashkina, O.I.; Anokhin, K.V.; Nikolaeva, A.Y.; Korzhenevskiy, D.A.; Vlaskina, A.V.; et al. Novel Genetically Encoded Bright Positive Calcium Indicator NCaMP7 Based on the mNeonGreen Fluorescent Protein. Int. J. Mol. Sci. 2020, 21, 1644. [Google Scholar] [CrossRef] [Green Version]
- Shyu, Y.J.; Liu, H.; Deng, X.; Hu, C.D. Identification of new fluorescent protein fragments for bimolecular fluorescence complementation analysis under physiological conditions. BioTechniques 2006, 40, 61–66. [Google Scholar] [CrossRef] [Green Version]
- Subach, O.M.; Barykina, N.V.; Anokhin, K.V.; Piatkevich, K.D.; Subach, F.V. Near-Infrared Genetically Encoded Positive Calcium Indicator Based on GAF-FP Bacterial Phytochrome. Int. J. Mol. Sci. 2019, 20, 3488. [Google Scholar] [CrossRef] [Green Version]
- Chen, T.W.; Wardill, T.J.; Sun, Y.; Pulver, S.R.; Renninger, S.L.; Baohan, A.; Schreiter, E.R.; Kerr, R.A.; Orger, M.B.; Jayaraman, V.; et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 2013, 499, 295–300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doronin, D.A.; Barykina, N.V.; Subach, O.M.; Sotskov, V.P.; Plusnin, V.V.; Ivleva, O.A.; Isaakova, E.A.; Varizhuk, A.M.; Pozmogova, G.E.; Malyshev, A.Y.; et al. Genetically encoded calcium indicator with NTnC-like design and enhanced fluorescence contrast and kinetics. BMC Biotechnol. 2018, 18, 10. [Google Scholar] [CrossRef] [PubMed]
- Moeyaert, B.; Holt, G.; Madangopal, R.; Perez-Alvarez, A.; Fearey, B.C.; Trojanowski, N.F.; Ledderose, J.; Zolnik, T.A.; Das, A.; Patel, D.; et al. Improved methods for marking active neuron populations. Nat. Commun. 2018, 9, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]







| Properties | FRCaMP | R-GECO1 a | ||||
|---|---|---|---|---|---|---|
| Apo | Sat | Apo | Sat | |||
| Absorption maximum (nm) | 446 (575) | 566 | 445 (577) | 561 | ||
| Excitation maximum (nm) | NF b (576) | 564 | NF * (578) * | 564 * | ||
| Emission maximum (nm) | NF (602) | 592 | NF * (600) | 589 | ||
| Quantum yield (%) c | <0.004 (0.133 ± 0.003) | 0.228 ± 0.008 | <0.006 * (0.06) | 0.20 | ||
| ε (mM−1·cm−1) d | 26.16 ± 0.08 (6.47 ± 0.07) | 53 ± 2 | 22 (15) | 51 | ||
| Brightness vs. EGFP (%) e | 0 (2.5) | 36 | 0 * (2.7) | 30 | ||
| pKa | 8.88 ± 0.05 | 6.60 ± 0.04 | 8.9 | 6.59 | ||
| ΔF/F | Purified protein | 0 mM Mg | 16.4 ± 0.7 | 20 ± 1 * | ||
| 1 mM Mg | 15.8 ± 0.5 | 21.0 ± 0.2 * | ||||
| HeLa cells | 5.6 ± 2.7 | 3.9 ± 1.7 * | ||||
| Kd (nM) f | 0 mM Mg | 214 ± 6 (2.5 ± 0.2) | 463 ± 10 (1.92 ± 0.07) * | |||
| 1 mM Mg | 441 ± 19 (2.7 ± 0.3) | 1138 ± 43 (2.02 ± 0.12) * | ||||
| Monomeric state | monomer | dimer | ||||
| Photobleaching halftime (sec) g | 4.3 ± 1.4 | 10.8 ± 5.3 | 28.5 ± 6.6 * | 25.6 ± 4.1 * | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Subach, O.M.; Barykina, N.V.; Chefanova, E.S.; Vlaskina, A.V.; Sotskov, V.P.; Ivashkina, O.I.; Anokhin, K.V.; Subach, F.V. FRCaMP, a Red Fluorescent Genetically Encoded Calcium Indicator Based on Calmodulin from Schizosaccharomyces Pombe Fungus. Int. J. Mol. Sci. 2021, 22, 111. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22010111
Subach OM, Barykina NV, Chefanova ES, Vlaskina AV, Sotskov VP, Ivashkina OI, Anokhin KV, Subach FV. FRCaMP, a Red Fluorescent Genetically Encoded Calcium Indicator Based on Calmodulin from Schizosaccharomyces Pombe Fungus. International Journal of Molecular Sciences. 2021; 22(1):111. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22010111
Chicago/Turabian StyleSubach, Oksana M., Natalia V. Barykina, Elizaveta S. Chefanova, Anna V. Vlaskina, Vladimir P. Sotskov, Olga I. Ivashkina, Konstantin V. Anokhin, and Fedor V. Subach. 2021. "FRCaMP, a Red Fluorescent Genetically Encoded Calcium Indicator Based on Calmodulin from Schizosaccharomyces Pombe Fungus" International Journal of Molecular Sciences 22, no. 1: 111. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22010111