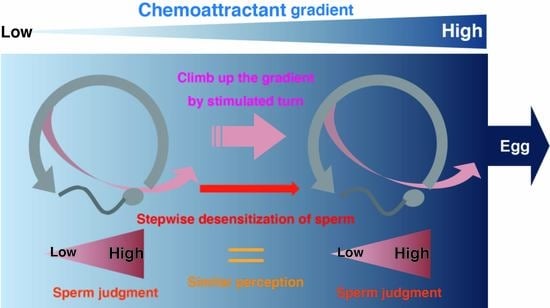
In Silico Reconstruction of Sperm Chemotaxis
Abstract
:1. Introduction
2. Results
2.1. Modeling of Sperm
2.2. Performance of Chemotaxis Model in Sea Urchin
2.3. Performance of Chemotaxis Model in Starfish
2.4. Further Analysis of Models
3. Discussion
4. Materials and Methods
Simulation Experiments
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ward, G.E.; Brokaw, C.J.; Garbers, D.L.; Vacquier, V.D. Chemotaxis of Arbacia punctulata spermatozoa to resact, a peptide from the egg jelly layer. J. Cell Biol. 1985, 101, 2324–2329. [Google Scholar] [CrossRef] [Green Version]
- Coll, J.C.; Bowden, B.F.; Meehan, G.V.; Konig, G.M.; Carroll, A.R.; Tapiolas, D.M.; Alino, P.M.; Heaton, A.; De Nys, R.; Leone, P.A.; et al. Chemical aspects of mass spawning in corals. I. Sperm-attractant molecules in the eggs of the scleractinian coral Montipora digitata. Mar. Biol. 1994, 118, 177–182. [Google Scholar] [CrossRef]
- Nishigaki, T.; Chiba, K.; Miki, W.; Hoshi, M. Structure and function of asterosaps, sperm-activating peptides from the jelly coat of starfish eggs. Zygote 1996, 4, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Olson, J.H.; Xiang, X.; Ziegert, T.; Kittelson, A.; Rawls, A.; Bieber, A.L.; Chandler, D.E. Allurin, a 21-kDa sperm chemoattractant from Xenopus egg jelly, is related to mammalian sperm-binding proteins. Proc. Natl. Acad. Sci. USA 2001, 98, 11205–11210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshida, M.; Murata, M.; Inaba, K.; Morisawa, M. A chemoattractant for ascidian spermatozoa is a sulfated steroid. Proc. Natl. Acad. Sci. USA 2002, 99, 14831–14836. [Google Scholar] [CrossRef] [Green Version]
- Teves, M.E.; Barbano, F.; Guidobaldi, H.A.; Sanchez, R.; Miska, W.; Giojalas, L.C. Progesterone at the picomolar range is a chemoattractant for mammalian spermatozoa. Fertil. Steril. 2006, 86, 745–749. [Google Scholar] [CrossRef]
- Oren-Benaroya, R.; Orvieto, R.; Gakamsky, A.; Pinchasov, M.; Eisenbach, M. The sperm chemoattractant secreted from human cumulus cells is progesterone. Hum. Reprod. 2008, 23, 2339–2345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strünker, T.; Alvarez, L.; Kaupp, U.B. At the physical limit—Chemosensation in sperm. Curr. Opin. Neurobiol. 2015, 34, 110–116. [Google Scholar] [CrossRef]
- Brenker, C.; Goodwin, N.; Weyand, I.; Kashikar, N.D.; Naruse, M.; Krahling, M.; Muller, A.; Kaupp, U.B.; Strünker, T. The CatSper channel: A polymodal chemosensor in human sperm. EMBO J. 2012, 31, 1654–1665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ernesto, J.I.; Weigel Muñoz, M.; Battistone, M.A.; Vasen, G.; Martínez-López, P.; Orta, G.; Figueiras-Fierro, D.; De la Vega-Beltran, J.L.; Moreno, I.A.; Guidobaldi, H.A.; et al. CRISP1 as a novel CatSper regulator that modulates sperm motility and orientation during fertilization. J. Cell Biol. 2015, 210, 1213–1224. [Google Scholar] [CrossRef]
- Nishigaki, T.; Chiba, K.; Hoshi, M. A 130-kDa membrane protein of sperm flagella is the receptor for asterosaps, sperm-activating peptides of starfish Asterias amurensis. Dev. Biol. 2000, 219, 154–162. [Google Scholar] [CrossRef] [Green Version]
- Singh, S.; Lowe, D.G.; Thorpe, D.S.; Rodriguez, H.; Kuang, W.-J.; Dangott, L.J.; Chinkers, M.; Goeddel, D.V.; Garbers, D.L. Membrane guanylate cyclase is a cell-surface receptor with homology to protein kinases. Nature 1988, 334, 708–712. [Google Scholar] [CrossRef] [PubMed]
- Brokaw, C.J. Calcium-induced asymmetrical beating of triton-demembranated sea urchin sperm flagella. J. Cell Biol. 1979, 82, 401–411. [Google Scholar] [CrossRef]
- Shiba, K.; Baba, S.A.; Inoue, T.; Yoshida, M. Ca2+ bursts occur around a local minimal concentration of attractant and trigger sperm chemotactic response. Proc. Natl. Acad. Sci. USA 2008, 105, 19312–19317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Böhmer, M.; Van, Q.; Weyland, I.; Hagen, V.; Beyermann, M.; Matsumoto, M.; Hoshi, M.; Hildebrand, E.; Kaupp, U.B. Ca2+ spikes in the flagellum control chemotactic behavior of sperm. EMBO J. 2005, 24, 2741–2752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaupp, U.B.; Solzin, J.; Hildebrand, E.; Brown, J.E.; Helbig, A.; Hagen, V.; Beyermann, M.; Pampaloni, F.; Weyand, I. The signal flow and motor response controling chemotaxis of sea urchin sperm. Nat. Cell Biol. 2003, 5, 109–117. [Google Scholar] [CrossRef]
- Matsumoto, M.; Kawase, O.; Islam, M.S.; Naruse, M.; Watanabe, S.-N.; Ishikawa, R.; Hoshi, M. Regulation of the starfish sperm acrosome reaction by cGMP, pH, cAMP and Ca2+. Int. J. Dev. Biol. 2008, 52, 523–526. [Google Scholar] [CrossRef] [Green Version]
- Bönigk, W.; Loogen, A.; Seifert, R.; Kashikar, N.; Klemm, C.; Krause, E.; Hagen, V.; Kremmer, E.; Strünker, T.; Kaupp, U.B. An atypical CNG channel activated by a single cGMP molecule controls sperm chemotaxis. Sci. Signal. 2009, 2, ra68. [Google Scholar] [CrossRef] [PubMed]
- Seifert, R.; Flick, M.; Bönigk, W.; Alvarez, L.; Trötschel, C.; Poetsch, A.; Muller, A.; Goodwin, N.; Pelzer, P.; Kashikar, N.D.; et al. The C at S per channel controls chemosensation in sea urchin sperm. EMBO J. 2015, 34, 379–392. [Google Scholar] [CrossRef] [Green Version]
- Trötschel, C.; Hamzeh, H.; Alvarez, L.; Pascal, R.; Lavryk, F.; Bönigk, W.; Körschen, H.G.; Müller, A.; Poetsch, A.; Rennhack, A.; et al. Absolute proteomic quantification reveals design principles of sperm flagellar chemosensation. EMBO J. 2020, 39, e102723. [Google Scholar] [CrossRef]
- Pichlo, M.; Bungert-Plümke, S.; Weyland, I.; Seifert, R.; Bönigk, W.; Strünker, T.; Kashikar, N.D.; Goodwin, N.; Müller, A.; Körschen, H.G.; et al. High density and ligand affinity confer ultrasensitive signal detection by a guanylyl cyclase chemoreceptor. J. Cell Biol. 2014, 206, 541–557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naruse, M.; Suetomo, H.; Matsubara, T.; Sato, T.; Yanagawa, H.; Hoshi, M.; Matsumoto, M. Acrosome reaction-related steroidal saponin, Co-ARIS, from the starfish induces structural changes in microdomains. Dev. Biol. 2010, 347, 147–153. [Google Scholar] [CrossRef] [Green Version]
- Niikura, K.; Alam, M.S.; Naruse, M.; Jimbo, M.; Moriyama, H.; Reich, A.; Wessel, G.M.; Matsumoto, M. Protein kinase A activity leads to the extension of the acrosomal process in starfish sperm. Mol. Reprod. Dev. 2017, 84, 614–625. [Google Scholar] [CrossRef] [PubMed]
- Ward, G.E.; Moy, G.W.; Vacquier, V.D. Dephosphorylation of sea urchin sperm guanylate cyclase during fertilization. In The Molecular and Cellular Biology of Fertilization; Springer: Boston, MA, USA, 1986; pp. 359–382. [Google Scholar] [CrossRef]
- Kawase, O.; Ueno, S.; Minakata, H.; Hoshi, M.; Matsumoto, M. Guanylyl cyclase and cGMP-specific phosphodiesterase participate in the acrosome reaction of starfish sperm. Zygote 2004, 12, 345–355. [Google Scholar] [CrossRef]
- Potter, L.R.; Garbers, D.L. Dephosphorylation of the guanylyl cyclase-A receptor causes desensitization. J. Biol. Chem. 1992, 267, 14531–14534. [Google Scholar] [CrossRef]
- Potter, L.R.; Hunter, T. Activation of protein kinase C stimulates the dephosphorylation of natriuretic peptide receptor-B at a single serine residue: A possible mechanism of heterologous desensitization. J. Biol. Chem. 2000, 275, 31099–31106. [Google Scholar] [CrossRef] [Green Version]
- Potter, L.R.; Hunter, T. Guanylyl cyclase-linked natriuretic peptide receptors: Structure and regulation. J. Biol. Chem. 2001, 276, 6057–6060. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Potthast, R.; Abbey-Hosch, S.E.; Antos, L.K.; Marchant, J.S.; Kuhn, M.; Potter, L.R. Calcium-dependent dephosphorylation mediates the hyperosmotic and lysophosphatidic acid-dependent inhibition of natriuretic peptide receptor-B/guanylyl cyclase-B. J. Biol. Chem. 2004, 279, 48513–48519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- MacKay, S.A. Computer simulation of aggregation in Dictyostelium discoideum. J. Cell Sci. 1978, 33, 1–16. [Google Scholar] [CrossRef]
- Tomlin, C.J.; Axelrod, J.D. Biology by numbers: Mathematical modeling in developmental biology. Nat. Rev. Genet. 2007, 8, 331–340. [Google Scholar] [CrossRef]
- Song, Y.; Cygnar, K.D.; Sagdullaev, B.; Valley, M.; Hirsh, S.; Stephan, A.; Reisert, J.; Zhao, H. Olfactory CNG channel desensitization by Ca2+/CaM via the B1b subunit affects response termination but not sensitivity to recurring stimulation. Neuron 2008, 58, 374–386. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, M.; Yoshida, K. Sperm chemotaxis and regulation of flagellar movement by Ca2+. Mol. Hum. Reprod. 2011, 17, 457–465. [Google Scholar] [CrossRef] [Green Version]
- Ramirez-Gómez, H.V.; Sabinina, V.J.; Pérez, M.V.; Beltran, C.; Carneiro, J.; Wood, C.D.; Tuval, I.; Darszon, A.; Guerrero, A. Sperm chemotaxis is driven by the slope of the chemoattractant concentration field. eLife 2020, 9, e50532. [Google Scholar] [CrossRef] [PubMed]
- Naruse, M.; Ishikawa, R.; Sakaya, H.; Moriyama, H.; Hoshi, M.; Matsumoto, M. Novel conserved structural domains of acrosome reaction-inducing substance are widespread in invertebrates. Mol. Reprod. Dev. 2011, 78, 57–66. [Google Scholar] [CrossRef]
- Kromer, J.A.; Märcker, S.; Lange, S.; Baier, C.; Friedrich, B.M. Decision making improves sperm chemotaxis in the presence of noise. PLoS Comput. Biol. 2018, 14, e1006109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guerrero, A.; Carneiro, J.; Pimentel, A.; Wood, C.D.; Corkidi, G.; Darszon, A. Strategies for locating the female gamete: The importance of measuring sperm trajectories in three spatial dimensions. Mol. Hum. Reprod. 2011, 17, 511–523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jikeli, J.F.; Alvarez, L.; Friedrich, B.M.; Wilson, L.G.; Pascal, R.; Colin, R.; Pichlo, M.; Rennhack, A.; Brenker, C.; Kaupp, U.B. Sperm navigation along helical paths in 3D chemoattractant landscapes. Nat. Commun. 2015, 6, 7985. [Google Scholar] [CrossRef] [Green Version]
- Alvarez, L.; Dai, L.; Friedrich, B.M.; Kashikar, N.D.; Gregor, I.; Pascal, R.; Kaupp, U.B. The rate of change in Ca2+ concentration controls sperm chemotaxis. J. Cell Biol. 2012, 196, 653–663. [Google Scholar] [CrossRef] [Green Version]
- Alvarez, L.; Friedrich, B.M.; Gompper, G.; Kaupp, U.B. The computational sperm cell. Trends Cell Biol. 2014, 24, 198–207. [Google Scholar] [CrossRef] [PubMed]
- Strünker, T.; Goodwin, N.; Brenker, C.; Kashikar, N.D.; Weyand, I.; Seifert, R.; Kaupp, U.B. The CatSper channel mediates progesterone-induced Ca2+ influx in human sperm. Nature 2011, 471, 382–386. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Naruse, M.; Matsumoto, M. In Silico Reconstruction of Sperm Chemotaxis. Int. J. Mol. Sci. 2021, 22, 9104. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22179104
Naruse M, Matsumoto M. In Silico Reconstruction of Sperm Chemotaxis. International Journal of Molecular Sciences. 2021; 22(17):9104. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22179104
Chicago/Turabian StyleNaruse, Masahiro, and Midori Matsumoto. 2021. "In Silico Reconstruction of Sperm Chemotaxis" International Journal of Molecular Sciences 22, no. 17: 9104. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22179104