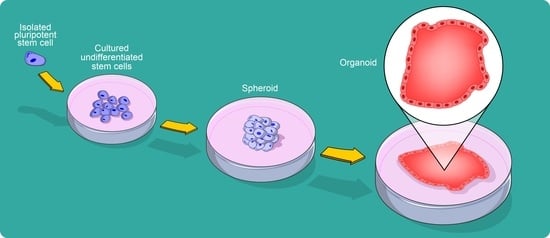
From Spheroids to Organoids: The Next Generation of Model Systems of Human Cardiac Regeneration in a Dish
Abstract
:1. Defining an Organoid
2. A Brief Historical Perspective of Organoid Development
3. The Basis of 3D Cellular Structure Formation
4. Organoid Generation
5. From Cardiac Spheroids to Cardioids
6. Cardioids
7. Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| aCM | adult cardiomyocyte |
| aSC | adult stem cell |
| CDC | cardiosphere-derived cell |
| CM | cardiomyocyte |
| COVID-19 | coronavirus disease 2019 |
| CSC | cardiac stem cell |
| EB | embryoid body |
| ECM | extracellular matrix |
| ESC | embryonic stem cell |
| hCO | human cardiac organoid |
| hiPSC | human induced pluripotent stem cell |
| hPSC | human pluripotent stem cell |
| iCM | contracting cardiomyocyte obtained from CSC-derived cardiosphere |
| iPSC | induced pluripotent stem cell |
| MSC | mesenchymal stem cell |
| PSC | pluripotent stem cell |
| SARS-CoV-2 | severe acute respiratory syndrome coronavirus 2 |
| SC | stem cell |
References
- Artegiani, B.; Clevers, H. Use and application of 3D-organoid technology. Hum. Mol. Genet. 2018, 27, R99–R107. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.; Koo, B.K.; Knoblich, J.A. Human organoids: Model systems for human biology and medicine. Nat. Rev. Mol. Cell Biol. 2020, 21, 571–584. [Google Scholar] [CrossRef]
- Fatehullah, A.; Tan, S.H.; Barker, N. Organoids as an in vitro model of human development and disease. Nat. Cell Biol. 2016, 18, 246–254. [Google Scholar] [CrossRef] [Green Version]
- Bahmad, H.F.; Elajami, M.K.; Daouk, R.; Jalloul, H.; Darwish, B.; Chalhoub, R.M.; Assi, S.; Chamaa, F.; Abou-Kheir, W. Stem Cells: In Sickness and in Health. Curr. Stem Cell Res. Ther. 2021, 16, 262–276. [Google Scholar] [CrossRef] [PubMed]
- Hubert, C.G.; Rivera, M.; Spangler, L.C.; Wu, Q.; Mack, S.C.; Prager, B.C.; Couce, M.; McLendon, R.E.; Sloan, A.E.; Rich, J.N. A Three-Dimensional Organoid Culture System Derived from Human Glioblastomas Recapitulates the Hypoxic Gradients and Cancer Stem Cell Heterogeneity of Tumors Found In Vivo. Cancer Res. 2016, 76, 2465–2477. [Google Scholar] [CrossRef] [Green Version]
- Wilson, H.V. A New Method by Which Sponges May Be Artificially Reared. Science 1907, 25, 912–915. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holtfreter, J. Experimental studies on the development of the pronephros. Rev. Can. Biol. 1944, 3, 220–250. [Google Scholar]
- Weiss, P.; Taylor, A.C. Reconstitution of Complete Organs from Single-Cell Suspensions of Chick Embryos in Advanced Stages of Differentiation. Proc. Natl. Acad. Sci. USA 1960, 46, 1177–1185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pierce, G.B.J.; Verney, E.L. An in vitro and in vivo study of differentiation in teratocarcinomas. Cancer 1961, 14, 1017–1029. [Google Scholar] [CrossRef]
- Steinberg, M.S. On the Mechanism of Tissue Reconstruction by Dissociated Cells, Iii. Free Energy Relations and the Reorganization of Fused, Heteronomic Tissue Fragments. Proc. Natl. Acad. Sci. USA 1962, 48, 1769–1776. [Google Scholar] [CrossRef] [Green Version]
- Evans, M.J.; Kaufman, M.H. Establishment in culture of pluripotential cells from mouse embryos. Nature 1981, 292, 154–156. [Google Scholar] [CrossRef] [PubMed]
- Martin, G.R. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl. Acad. Sci. USA 1981, 78, 7634–7638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, M.L.; Aggeler, J.; Farson, D.A.; Hatier, C.; Hassell, J.; Bissell, M.J. Influence of a reconstituted basement membrane and its components on casein gene expression and secretion in mouse mammary epithelial cells. Proc. Natl. Acad. Sci. USA 1987, 84, 136–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shannon, J.M.; Mason, R.J.; Jennings, S.D. Functional differentiation of alveolar type II epithelial cells in vitro: Effects of cell shape, cell-matrix interactions and cell-cell interactions. Biochim. Biophys. Acta 1987, 931, 143–156. [Google Scholar] [CrossRef]
- Thomson, J.A.; Itskovitz-Eldor, J.; Shapiro, S.S.; Waknitz, M.A.; Swiergiel, J.J.; Marshall, V.S.; Jones, J.M. Embryonic stem cell lines derived from human blastocysts. Science 1998, 282, 1145–1147. [Google Scholar] [CrossRef] [Green Version]
- Eiraku, M.; Watanabe, K.; Matsuo-Takasaki, M.; Kawada, M.; Yonemura, S.; Matsumura, M.; Wataya, T.; Nishiyama, A.; Muguruma, K.; Sasai, Y. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell 2008, 3, 519–532. [Google Scholar] [CrossRef] [Green Version]
- Sato, T.; Vries, R.G.; Snippert, H.J.; van de Wetering, M.; Barker, N.; Stange, D.E.; van Es, J.H.; Abo, A.; Kujala, P.; Peters, P.J.; et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 2009, 459, 262–265. [Google Scholar] [CrossRef]
- Spence, J.R.; Mayhew, C.N.; Rankin, S.A.; Kuhar, M.F.; Vallance, J.E.; Tolle, K.; Hoskins, E.E.; Kalinichenko, V.V.; Wells, S.I.; Zorn, A.M.; et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature 2011, 470, 105–109. [Google Scholar] [CrossRef] [Green Version]
- Eiraku, M.; Sasai, Y. Mouse embryonic stem cell culture for generation of three-dimensional retinal and cortical tissues. Nat. Protoc. 2011, 7, 69–79. [Google Scholar] [CrossRef]
- Lancaster, M.A.; Renner, M.; Martin, C.A.; Wenzel, D.; Bicknell, L.S.; Hurles, M.E.; Homfray, T.; Penninger, J.M.; Jackson, A.P.; Knoblich, J.A. Cerebral organoids model human brain development and microcephaly. Nature 2013, 501, 373–379. [Google Scholar] [CrossRef]
- Takasato, M.; Er, P.X.; Chiu, H.S.; Maier, B.; Baillie, G.J.; Ferguson, C.; Parton, R.G.; Wolvetang, E.J.; Roost, M.S.; Chuva de Sousa Lopes, S.M.; et al. Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature 2015, 526, 564–568. [Google Scholar] [CrossRef] [PubMed]
- Low, J.H.; Li, P.; Chew, E.G.Y.; Zhou, B.; Suzuki, K.; Zhang, T.; Lian, M.M.; Liu, M.; Aizawa, E.; Rodriguez Esteban, C.; et al. Generation of Human PSC-Derived Kidney Organoids with Patterned Nephron Segments and a De Novo Vascular Network. Cell Stem Cell 2019, 25, 373–387.e9. [Google Scholar] [CrossRef] [PubMed]
- Prior, N.; Inacio, P.; Huch, M. Liver organoids: From basic research to therapeutic applications. Gut 2019, 68, 2228–2237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balak, J.R.A.; Juksar, J.; Carlotti, F.; Lo Nigro, A.; de Koning, E.J.P. Organoids from the Human Fetal and Adult Pancreas. Curr. Diabetes Rep. 2019, 19, 160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takayama, K. In Vitro and Animal Models for SARS-CoV-2 research. Trends Pharmacol. Sci. 2020, 41, 513–517. [Google Scholar] [CrossRef]
- Lamers, M.M.; van der Vaart, J.; Knoops, K.; Riesebosch, S.; Breugem, T.I.; Mykytyn, A.Z.; Beumer, J.; Schipper, D.; Bezstarosti, K.; Koopman, C.D.; et al. An organoid-derived bronchioalveolar model for SARS-CoV-2 infection of human alveolar type II-like cells. EMBO J. 2021, 40, e105912. [Google Scholar] [CrossRef]
- Wimmer, R.A.; Leopoldi, A.; Aichinger, M.; Wick, N.; Hantusch, B.; Novatchkova, M.; Taubenschmid, J.; Hämmerle, M.; Esk, C.; Bagley, J.A.; et al. Human blood vessel organoids as a model of diabetic vasculopathy. Nature 2019, 565, 505–510. [Google Scholar] [CrossRef]
- Hofbauer, P.; Jahnel, S.M.; Papai, N.; Giesshammer, M.; Deyett, A.; Schmidt, C.; Penc, M.; Tavernini, K.; Grdseloff, N.; Meledeth, C.; et al. Cardioids reveal self-organizing principles of human cardiogenesis. Cell 2021, 184, 3299–3317.e22. [Google Scholar] [CrossRef]
- Armstrong, P.B. Cell sorting out: The self-assembly of tissues in vitro. Crit. Rev. Biochem. Mol. Biol. 1989, 24, 119–149. [Google Scholar] [CrossRef]
- Steinberg, M.S.; Roth, S.A. Phases in Cell Aggregation and Tissue Reconstruction an Approach to the Kinetics of Cell Aggregation. J. Exp. Zool. 1964, 157, 327–338. [Google Scholar] [CrossRef]
- Fujimori, T.; Miyatani, S.; Takeichi, M. Ectopic expression of N-cadherin perturbs histogenesis in Xenopus embryos. Development 1990, 110, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Detrick, R.J.; Dickey, D.; Kintner, C.R. The effects of N-cadherin misexpression on morphogenesis in Xenopus embryos. Neuron 1990, 4, 493–506. [Google Scholar] [CrossRef]
- Agathocleous, M.; Harris, W.A. From progenitors to differentiated cells in the vertebrate retina. Annu. Rev. Cell Dev. Biol. 2009, 25, 45–69. [Google Scholar] [CrossRef]
- Zigman, M.; Cayouette, M.; Charalambous, C.; Schleiffer, A.; Hoeller, O.; Dunican, D.; McCudden, C.R.; Firnberg, N.; Barres, B.A.; Siderovski, D.P.; et al. Mammalian inscuteable regulates spindle orientation and cell fate in the developing retina. Neuron 2005, 48, 539–545. [Google Scholar] [CrossRef] [Green Version]
- Moscona, A. Rotation-mediated histogenetic aggregation of dissociated cells. A quantifiable approach to cell interactions in vitro. Exp. Cell Res. 1961, 22, 455–475. [Google Scholar] [CrossRef]
- Moscona, A.; Moscona, H. The dissociation and aggregation of cells from organ rudiments of the early chick embryo. J. Anat. 1952, 86, 287–301. [Google Scholar] [PubMed]
- Mallon, B.S.; Park, K.Y.; Chen, K.G.; Hamilton, R.S.; McKay, R.D. Toward xeno-free culture of human embryonic stem cells. Int. J. Biochem. Cell Biol. 2006, 38, 1063–1075. [Google Scholar] [CrossRef] [Green Version]
- Pampaloni, F.; Reynaud, E.G.; Stelzer, E.H. The third dimension bridges the gap between cell culture and live tissue. Nat. Rev. Mol. Cell Biol. 2007, 8, 839–845. [Google Scholar] [CrossRef]
- Breslin, S.; O’Driscoll, L. Three-dimensional cell culture: The missing link in drug discovery. Drug Discov. Today 2013, 18, 240–249. [Google Scholar] [CrossRef]
- Guilak, F.; Cohen, D.M.; Estes, B.T.; Gimble, J.M.; Liedtke, W.; Chen, C.S. Control of stem cell fate by physical interactions with the extracellular matrix. Cell Stem Cell 2009, 5, 17–26. [Google Scholar] [CrossRef] [Green Version]
- Knight, E.; Przyborski, S. Advances in 3D cell culture technologies enabling tissue-like structures to be created in vitro. J. Anat. 2015, 227, 746–756. [Google Scholar] [CrossRef] [Green Version]
- Thomas, C.H.; Collier, J.H.; Sfeir, C.S.; Healy, K.E. Engineering gene expression and protein synthesis by modulation of nuclear shape. Proc. Natl. Acad. Sci. USA 2002, 99, 1972–1977. [Google Scholar] [CrossRef] [Green Version]
- Vicinanza, C.; Aquila, I.; Scalise, M.; Cristiano, F.; Marino, F.; Cianflone, E.; Mancuso, T.; Marotta, P.; Sacco, W.; Lewis, F.C.; et al. Adult cardiac stem cells are multipotent and robustly myogenic: C-kit expression is necessary but not sufficient for their identification. Cell Death Differ. 2017, 24, 2101–2116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kilian, K.A.; Bugarija, B.; Lahn, B.T.; Mrksich, M. Geometric cues for directing the differentiation of mesenchymal stem cells. Proc. Natl. Acad. Sci. USA 2010, 107, 4872–4877. [Google Scholar] [CrossRef] [Green Version]
- Nelson, C.M.; Bissell, M.J. Of extracellular matrix, scaffolds, and signaling: Tissue architecture regulates development, homeostasis, and cancer. Annu. Rev. Cell Dev. Biol. 2006, 22, 287–309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gattazzo, F.; Urciuolo, A.; Bonaldo, P. Extracellular matrix: A dynamic microenvironment for stem cell niche. Biochim. Biophys. Acta 2014, 1840, 2506–2519. [Google Scholar] [CrossRef] [PubMed]
- Skardal, A.; Shupe, T.; Atala, A. Organoid-on-a-chip and body-on-a-chip systems for drug screening and disease modeling. Drug Discov. Today 2016, 21, 1399–1411. [Google Scholar] [CrossRef] [PubMed]
- Baker, B.M.; Chen, C.S. Deconstructing the third dimension: How 3D culture microenvironments alter cellular cues. J. Cell Sci. 2012, 125, 3015–3024. [Google Scholar] [CrossRef] [Green Version]
- Yamamoto, H.; Quinn, G.; Asari, A.; Yamanokuchi, H.; Teratani, T.; Terada, M.; Ochiya, T. Differentiation of embryonic stem cells into hepatocytes: Biological functions and therapeutic application. Hepatology 2003, 37, 983–993. [Google Scholar] [CrossRef]
- Ishii, T.; Fukumitsu, K.; Yasuchika, K.; Adachi, K.; Kawase, E.; Suemori, H.; Nakatsuji, N.; Ikai, I.; Uemoto, S. Effects of extracellular matrixes and growth factors on the hepatic differentiation of human embryonic stem cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2008, 295, G313–G321. [Google Scholar] [CrossRef] [Green Version]
- Hwang, N.S.; Varghese, S.; Elisseeff, J. Controlled differentiation of stem cells. Adv. Drug Deliv. Rev. 2008, 60, 199–214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haycock, J.W. 3D cell culture: A review of current approaches and techniques. Methods Mol. Biol. 2011, 695, 1–15. [Google Scholar] [CrossRef]
- Bhadriraju, K.; Chen, C.S. Engineering cellular microenvironments to improve cell-based drug testing. Drug Discov. Today 2002, 7, 612–620. [Google Scholar] [CrossRef]
- Jozefczuk, J.; Drews, K.; Adjaye, J. Preparation of mouse embryonic fibroblast cells suitable for culturing human embryonic and induced pluripotent stem cells. J. Vis. Exp. 2012, 64. [Google Scholar] [CrossRef] [Green Version]
- Chen, G.; Gulbranson, D.R.; Hou, Z.; Bolin, J.M.; Ruotti, V.; Probasco, M.D.; Smuga-Otto, K.; Howden, S.E.; Diol, N.R.; Propson, N.E.; et al. Chemically defined conditions for human iPSC derivation and culture. Nat. Methods 2011, 8, 424–429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Villa-Diaz, L.G.; Ross, A.M.; Lahann, J.; Krebsbach, P.H. Concise review: The evolution of human pluripotent stem cell culture: From feeder cells to synthetic coatings. Stem Cells 2013, 31, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Baraniak, P.R.; McDevitt, T.C. Scaffold-free culture of mesenchymal stem cell spheroids in suspension preserves multilineage potential. Cell Tissue Res. 2012, 347, 701–711. [Google Scholar] [CrossRef] [Green Version]
- Rosso, F.; Giordano, A.; Barbarisi, M.; Barbarisi, A. From cell-ECM interactions to tissue engineering. J. Cell Physiol. 2004, 199, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Keller, G.M. In vitro differentiation of embryonic stem cells. Curr. Opin. Cell Biol. 1995, 7, 862–869. [Google Scholar] [CrossRef]
- Wang, Y.; Kim, M.H.; Tabaei, S.R.; Park, J.H.; Na, K.; Chung, S.; Zhdanov, V.P.; Cho, N.J. Spheroid Formation of Hepatocarcinoma Cells in Microwells: Experiments and Monte Carlo Simulations. PLoS ONE 2016, 11, e0161915. [Google Scholar] [CrossRef] [Green Version]
- Lin, R.Z.; Chang, H.Y. Recent advances in three-dimensional multicellular spheroid culture for biomedical research. Biotechnol. J. 2008, 3, 1172–1184. [Google Scholar] [CrossRef]
- Cui, X.; Hartanto, Y.; Zhang, H. Advances in multicellular spheroids formation. J. R. Soc. Interface 2017, 14. [Google Scholar] [CrossRef] [PubMed]
- Edmondson, R.; Broglie, J.J.; Adcock, A.F.; Yang, L. Three-dimensional cell culture systems and their applications in drug discovery and cell-based biosensors. Assay Drug Dev. Technol. 2014, 12, 207–218. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Itaka, K.; Ohba, S.; Nishiyama, N.; Chung, U.I.; Yamasaki, Y.; Kataoka, K. 3D spheroid culture system on micropatterned substrates for improved differentiation efficiency of multipotent mesenchymal stem cells. Biomaterials 2009, 30, 2705–2715. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Guo, G.; Li, L.; Chen, F.; Bao, J.; Shi, Y.J.; Bu, H. Three-dimensional spheroid culture of human umbilical cord mesenchymal stem cells promotes cell yield and stemness maintenance. Cell Tissue Res. 2015, 360, 297–307. [Google Scholar] [CrossRef]
- Guo, H.; Ingolia, N.T.; Weissman, J.S.; Bartel, D.P. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 2010, 466, 835–840. [Google Scholar] [CrossRef] [Green Version]
- Shankaran, A.; Prasad, K.; Chaudhari, S.; Brand, A.; Satyamoorthy, K. Advances in development and application of human organoids. 3 Biotech 2021, 11, 257. [Google Scholar] [CrossRef]
- McCracken, K.W.; Cata, E.M.; Crawford, C.M.; Sinagoga, K.L.; Schumacher, M.; Rockich, B.E.; Tsai, Y.H.; Mayhew, C.N.; Spence, J.R.; Zavros, Y.; et al. Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature 2014, 516, 400–404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Nadauld, L.; Ootani, A.; Corney, D.C.; Pai, R.K.; Gevaert, O.; Cantrell, M.A.; Rack, P.G.; Neal, J.T.; Chan, C.W.; et al. Oncogenic transformation of diverse gastrointestinal tissues in primary organoid culture. Nat. Med. 2014, 20, 769–777. [Google Scholar] [CrossRef] [Green Version]
- Drost, J.; van Jaarsveld, R.H.; Ponsioen, B.; Zimberlin, C.; van Boxtel, R.; Buijs, A.; Sachs, N.; Overmeer, R.M.; Offerhaus, G.J.; Begthel, H.; et al. Sequential cancer mutations in cultured human intestinal stem cells. Nature 2015, 521, 43–47. [Google Scholar] [CrossRef]
- Matano, M.; Date, S.; Shimokawa, M.; Takano, A.; Fujii, M.; Ohta, Y.; Watanabe, T.; Kanai, T.; Sato, T. Modeling colorectal cancer using CRISPR-Cas9–mediated engineering of human intestinal organoids. Nat. Med. 2015, 21, 256–262. [Google Scholar] [CrossRef] [PubMed]
- Reubinoff, B.E.; Pera, M.F.; Fong, C.Y.; Trounson, A.; Bongso, A. Embryonic stem cell lines from human blastocysts: Somatic differentiation in vitro. Nat. Biotechnol. 2000, 18, 399–404. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [Green Version]
- Lancaster, M.A.; Knoblich, J.A. Organogenesis in a dish: Modeling development and disease using organoid technologies. Science 2014, 345, 1247125. [Google Scholar] [CrossRef]
- D’Amour, K.A.; Agulnick, A.D.; Eliazer, S.; Kelly, O.G.; Kroon, E.; Baetge, E.E. Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat. Biotechnol. 2005, 23, 1534–1541. [Google Scholar] [CrossRef]
- Kubo, A.; Shinozaki, K.; Shannon, J.M.; Kouskoff, V.; Kennedy, M.; Woo, S.; Fehling, H.J.; Keller, G. Development of definitive endoderm from embryonic stem cells in culture. Development 2004, 131, 1651–1662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mansour, A.A.; Goncalves, J.T.; Bloyd, C.W.; Li, H.; Fernandes, S.; Quang, D.; Johnston, S.; Parylak, S.L.; Jin, X.; Gage, F.H. An in vivo model of functional and vascularized human brain organoids. Nat. Biotechnol. 2018, 36, 432–441. [Google Scholar] [CrossRef]
- Rheinwald, J.G.; Green, H. Serial cultivation of strains of human epidermal keratinocytes: The formation of keratinizing colonies from single cells. Cell 1975, 6, 331–343. [Google Scholar] [CrossRef]
- McCauley, H.A.; Wells, J.M. Pluripotent stem cell-derived organoids: Using principles of developmental biology to grow human tissues in a dish. Development 2017, 144, 958–962. [Google Scholar] [CrossRef] [Green Version]
- Lancaster, M.A.; Huch, M. Disease modelling in human organoids. Dis. Models Mech. 2019, 12, dmm039347. [Google Scholar] [CrossRef] [Green Version]
- Xu, C.; Inokuma, M.S.; Denham, J.; Golds, K.; Kundu, P.; Gold, J.D.; Carpenter, M.K. Feeder-free growth of undifferentiated human embryonic stem cells. Nat. Biotechnol. 2001, 19, 971–974. [Google Scholar] [CrossRef]
- Kleinman, H.K.; Martin, G.R. Matrigel: Basement membrane matrix with biological activity. Semin. Cancer Biol. 2005, 15, 378–386. [Google Scholar] [CrossRef]
- Aisenbrey, E.A.; Murphy, W.L. Synthetic alternatives to Matrigel. Nat. Rev. Mater. 2020, 5, 539–551. [Google Scholar] [CrossRef] [PubMed]
- Broguiere, N.; Isenmann, L.; Hirt, C.; Ringel, T.; Placzek, S.; Cavalli, E.; Ringnalda, F.; Villiger, L.; Zullig, R.; Lehmann, R.; et al. Growth of Epithelial Organoids in a Defined Hydrogel. Adv. Mater. 2018, 30, e1801621. [Google Scholar] [CrossRef] [PubMed]
- Jabaji, Z.; Brinkley, G.J.; Khalil, H.A.; Sears, C.M.; Lei, N.Y.; Lewis, M.; Stelzner, M.; Martin, M.G.; Dunn, J.C. Type I collagen as an extracellular matrix for the in vitro growth of human small intestinal epithelium. PLoS ONE 2014, 9, e107814. [Google Scholar] [CrossRef] [Green Version]
- Lindborg, B.A.; Brekke, J.H.; Vegoe, A.L.; Ulrich, C.B.; Haider, K.T.; Subramaniam, S.; Venhuizen, S.L.; Eide, C.R.; Orchard, P.J.; Chen, W.; et al. Rapid Induction of Cerebral Organoids From Human Induced Pluripotent Stem Cells Using a Chemically Defined Hydrogel and Defined Cell Culture Medium. Stem Cells Transl. Med. 2016, 5, 970–979. [Google Scholar] [CrossRef] [Green Version]
- Gjorevski, N.; Sachs, N.; Manfrin, A.; Giger, S.; Bragina, M.E.; Ordonez-Moran, P.; Clevers, H.; Lutolf, M.P. Designer matrices for intestinal stem cell and organoid culture. Nature 2016, 539, 560–564. [Google Scholar] [CrossRef]
- Magin, C.M.; Alge, D.L.; Anseth, K.S. Bio-inspired 3D microenvironments: A new dimension in tissue engineering. Biomed. Mater. 2016, 11, 022001. [Google Scholar] [CrossRef] [PubMed]
- Engler, A.J.; Sen, S.; Sweeney, H.L.; Discher, D.E. Matrix elasticity directs stem cell lineage specification. Cell 2006, 126, 677–689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bateman, J.F.; Boot-Handford, R.P.; Lamande, S.R. Genetic diseases of connective tissues: Cellular and extracellular effects of ECM mutations. Nat. Rev. Genet. 2009, 10, 173–183. [Google Scholar] [CrossRef]
- Cox, T.R.; Erler, J.T. Remodeling and homeostasis of the extracellular matrix: Implications for fibrotic diseases and cancer. Dis. Model. Mech. 2011, 4, 165–178. [Google Scholar] [CrossRef] [Green Version]
- Timmins, N.E.; Nielsen, L.K. Generation of multicellular tumor spheroids by the hanging-drop method. Methods Mol. Med. 2007, 140, 141–151. [Google Scholar] [CrossRef] [PubMed]
- Kalabis, J.; Wong, G.S.; Vega, M.E.; Natsuizaka, M.; Robertson, E.S.; Herlyn, M.; Nakagawa, H.; Rustgi, A.K. Isolation and characterization of mouse and human esophageal epithelial cells in 3D organotypic culture. Nat. Protoc. 2012, 7, 235–246. [Google Scholar] [CrossRef] [Green Version]
- Turner, D.A.; Baillie-Johnson, P.; Martinez Arias, A. Organoids and the genetically encoded self-assembly of embryonic stem cells. Bioessays 2016, 38, 181–191. [Google Scholar] [CrossRef] [Green Version]
- Brassard, J.A.; Lutolf, M.P. Engineering Stem Cell Self-organization to Build Better Organoids. Cell Stem Cell 2019, 24, 860–876. [Google Scholar] [CrossRef] [PubMed]
- Fujimichi, Y.; Otsuka, K.; Tomita, M.; Iwasaki, T. An Efficient Intestinal Organoid System of Direct Sorting to Evaluate Stem Cell Competition in Vitro. Sci. Rep. 2019, 9, 20297. [Google Scholar] [CrossRef] [Green Version]
- Bauwens, C.L.; Peerani, R.; Niebruegge, S.; Woodhouse, K.A.; Kumacheva, E.; Husain, M.; Zandstra, P.W. Control of human embryonic stem cell colony and aggregate size heterogeneity influences differentiation trajectories. Stem Cells 2008, 26, 2300–2310. [Google Scholar] [CrossRef]
- Amaral, A.J.R.; Pasparakis, G. Cell membrane engineering with synthetic materials: Applications in cell spheroids, cellular glues and microtissue formation. Acta Biomater. 2019, 90, 21–36. [Google Scholar] [CrossRef] [PubMed]
- Rogozhnikov, D.; O’Brien, P.J.; Elahipanah, S.; Yousaf, M.N. Scaffold Free Bio-orthogonal Assembly of 3-Dimensional Cardiac Tissue via Cell Surface Engineering. Sci Rep. 2016, 6, 39806. [Google Scholar] [CrossRef] [Green Version]
- Luo, W.; Pulsipher, A.; Dutta, D.; Lamb, B.M.; Yousaf, M.N. Remote control of tissue interactions via engineered photo-switchable cell surfaces. Sci. Rep. 2014, 4, 6313. [Google Scholar] [CrossRef] [Green Version]
- Akbari, E.; Mollica, M.Y.; Lucas, C.R.; Bushman, S.M.; Patton, R.A.; Shahhosseini, M.; Song, J.W.; Castro, C.E. Engineering Cell Surface Function with DNA Origami. Adv. Mater. 2017, 29, 1703632. [Google Scholar] [CrossRef]
- Fayol, D.; Frasca, G.; Le Visage, C.; Gazeau, F.; Luciani, N.; Wilhelm, C. Use of magnetic forces to promote stem cell aggregation during differentiation, and cartilage tissue modeling. Adv. Mater. 2013, 25, 2611–2616. [Google Scholar] [CrossRef]
- Xia, P.F.; Ling, H.; Foo, J.L.; Chang, M.W. Synthetic genetic circuits for programmable biological functionalities. Biotechnol. Adv. 2019, 37, 107393. [Google Scholar] [CrossRef] [PubMed]
- Bashor, C.J.; Collins, J.J. Understanding Biological Regulation Through Synthetic Biology. Annu. Rev. Biophys. 2018, 47, 399–423. [Google Scholar] [CrossRef]
- Gaj, T.; Gersbach, C.A.; Barbas, C.F., III. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013, 31, 397–405. [Google Scholar] [CrossRef] [Green Version]
- Schwank, G.; Koo, B.K.; Sasselli, V.; Dekkers, J.F.; Heo, I.; Demircan, T.; Sasaki, N.; Boymans, S.; Cuppen, E.; van der Ent, C.K.; et al. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell 2013, 13, 653–658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Driehuis, E.; Clevers, H. CRISPR/Cas 9 genome editing and its applications in organoids. Am. J. Physiol. Gastrointest. Liver Physiol. 2017, 312, G257–G265. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.L.; Gao, M.L.; Lei, X.L.; Lv, J.N.; Zhao, H.; He, K.W.; Xia, X.X.; Li, L.Y.; Chen, Y.C.; Li, Y.P.; et al. Gene Correction Reverses Ciliopathy and Photoreceptor Loss in iPSC-Derived Retinal Organoids from Retinitis Pigmentosa Patients. Stem Cell Rep. 2018, 10, 1267–1281. [Google Scholar] [CrossRef] [Green Version]
- Freedman, B.S.; Brooks, C.R.; Lam, A.Q.; Fu, H.; Morizane, R.; Agrawal, V.; Saad, A.F.; Li, M.K.; Hughes, M.R.; Werff, R.V.; et al. Modelling kidney disease with CRISPR-mutant kidney organoids derived from human pluripotent epiblast spheroids. Nat. Commun. 2015, 6, 8715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gracz, A.D.; Williamson, I.A.; Roche, K.C.; Johnston, M.J.; Wang, F.; Wang, Y.; Attayek, P.J.; Balowski, J.; Liu, X.F.; Laurenza, R.J.; et al. A high-throughput platform for stem cell niche co-cultures and downstream gene expression analysis. Nat. Cell Biol. 2015, 17, 340–349. [Google Scholar] [CrossRef] [Green Version]
- Allazetta, S.; Lutolf, M.P. Stem cell niche engineering through droplet microfluidics. Curr. Opin. Biotechnol. 2015, 35, 86–93. [Google Scholar] [CrossRef]
- Murphy, S.V.; Atala, A. 3D bioprinting of tissues and organs. Nat. Biotechnol. 2014, 32, 773–785. [Google Scholar] [CrossRef]
- Wang, Y.; Gunasekara, D.B.; Reed, M.I.; DiSalvo, M.; Bultman, S.J.; Sims, C.E.; Magness, S.T.; Allbritton, N.L. A microengineered collagen scaffold for generating a polarized crypt-villus architecture of human small intestinal epithelium. Biomaterials 2017, 128, 44–55. [Google Scholar] [CrossRef] [PubMed]
- Nikolaev, M.; Mitrofanova, O.; Broguiere, N.; Geraldo, S.; Dutta, D.; Tabata, Y.; Elci, B.; Brandenberg, N.; Kolotuev, I.; Gjorevski, N.; et al. Homeostatic mini-intestines through scaffold-guided organoid morphogenesis. Nature 2020, 585, 574–578. [Google Scholar] [CrossRef] [PubMed]
- Kloxin, A.M.; Kasko, A.M.; Salinas, C.N.; Anseth, K.S. Photodegradable hydrogels for dynamic tuning of physical and chemical properties. Science 2009, 324, 59–63. [Google Scholar] [CrossRef] [Green Version]
- Mosiewicz, K.A.; Kolb, L.; van der Vlies, A.J.; Martino, M.M.; Lienemann, P.S.; Hubbell, J.A.; Ehrbar, M.; Lutolf, M.P. In situ cell manipulation through enzymatic hydrogel photopatterning. Nat. Mater. 2013, 12, 1072–1078. [Google Scholar] [CrossRef] [PubMed]
- Ruskowitz, E.R.; DeForest, C.A. Photoresponsive biomaterials for targeted drug delivery and 4D cell culture. Nat. Rev. Mater. 2018, 3, 17087. [Google Scholar] [CrossRef]
- Demers, C.J.; Soundararajan, P.; Chennampally, P.; Cox, G.A.; Briscoe, J.; Collins, S.D.; Smith, R.L. Development-on-chip: In vitro neural tube patterning with a microfluidic device. Development 2016, 143, 1884–1892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wylie, R.G.; Ahsan, S.; Aizawa, Y.; Maxwell, K.L.; Morshead, C.M.; Shoichet, M.S. Spatially controlled simultaneous patterning of multiple growth factors in three-dimensional hydrogels. Nat. Mater. 2011, 10, 799–806. [Google Scholar] [CrossRef] [PubMed]
- Broguiere, N.; Luchtefeld, I.; Trachsel, L.; Mazunin, D.; Rizzo, R.; Bode, J.W.; Lutolf, M.P.; Zenobi-Wong, M. Morphogenesis Guided by 3D Patterning of Growth Factors in Biological Matrices. Adv. Mater. 2020, 32, e1908299. [Google Scholar] [CrossRef]
- Torgersen, J.; Qin, X.H.; Li, Z.; Ovsianikov, A.; Liska, R.; Stampfl, J. Hydrogels for Two-Photon Polymerization: A Toolbox for Mimicking the Extracellular Matrix. Adv. Funct. Mat. 2013, 23, 4542–4554. [Google Scholar] [CrossRef] [Green Version]
- Matsa, E.; Burridge, P.W.; Wu, J.C. Human stem cells for modeling heart disease and for drug discovery. Sci. Transl. Med. 2014, 6, 239ps6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, D.C.; Hookway, T.A.; Wu, Q.; Jha, R.; Preininger, M.K.; Chen, X.; Easley, C.A.; Spearman, P.; Deshpande, S.R.; Maher, K.; et al. Microscale generation of cardiospheres promotes robust enrichment of cardiomyocytes derived from human pluripotent stem cells. Stem Cell Rep. 2014, 3, 260–268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kattman, S.J.; Witty, A.D.; Gagliardi, M.; Dubois, N.C.; Niapour, M.; Hotta, A.; Ellis, J.; Keller, G. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell 2011, 8, 228–240. [Google Scholar] [CrossRef] [Green Version]
- Rajala, K.; Pekkanen-Mattila, M.; Aalto-Setälä, K. Cardiac differentiation of pluripotent stem cells. Stem Cells Int. 2011, 2011, 383709. [Google Scholar] [CrossRef] [Green Version]
- Kempf, H.; Olmer, R.; Kropp, C.; Rückert, M.; Jara-Avaca, M.; Robles-Diaz, D.; Franke, A.; Elliott, D.A.; Wojciechowski, D.; Fischer, M.; et al. Controlling expansion and cardiomyogenic differentiation of human pluripotent stem cells in scalable suspension culture. Stem Cell Rep. 2014, 3, 1132–1146. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Soonpaa, M.H.; Adler, E.D.; Roepke, T.K.; Kattman, S.J.; Kennedy, M.; Henckaerts, E.; Bonham, K.; Abbott, G.W.; Linden, R.M.; et al. Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population. Nature 2008, 453, 524–528. [Google Scholar] [CrossRef]
- Elliott, D.A.; Braam, S.R.; Koutsis, K.; Ng, E.S.; Jenny, R.; Lagerqvist, E.L.; Biben, C.; Hatzistavrou, T.; Hirst, C.E.; Yu, Q.C.; et al. NKX2-5(eGFP/w) hESCs for isolation of human cardiac progenitors and cardiomyocytes. Nat. Methods 2011, 8, 1037–1040. [Google Scholar] [CrossRef]
- Yan, Y.; Bejoy, J.; Xia, J.; Griffin, K.; Guan, J.; Li, Y. Cell population balance of cardiovascular spheroids derived from human induced pluripotent stem cells. Sci. Rep. 2019, 9, 1295. [Google Scholar] [CrossRef] [PubMed]
- Correia, C.; Koshkin, A.; Duarte, P.; Hu, D.; Carido, M.; Sebastião, M.J.; Gomes-Alves, P.; Elliott, D.A.; Domian, I.J.; Teixeira, A.P.; et al. 3D aggregate culture improves metabolic maturation of human pluripotent stem cell derived cardiomyocytes. Biotechnol. Bioeng. 2018, 115, 630–644. [Google Scholar] [CrossRef]
- Gherghiceanu, M.; Barad, L.; Novak, A.; Reiter, I.; Itskovitz-Eldor, J.; Binah, O.; Popescu, L.M. Cardiomyocytes derived from human embryonic and induced pluripotent stem cells: Comparative ultrastructure. J. Cell. Mol. Med. 2011, 15, 2539–2551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, Y.F.; Chen, M.; Zhang, N.N.; Yang, H.J.; Rui, Q.; Zhou, Y.F. In vitro and in vivo differentiation of induced pluripotent stem cells generated from urine-derived cells into cardiomyocytes. Biol. Open 2018, 7, bio029157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Devalla, H.D.; Schwach, V.; Ford, J.W.; Milnes, J.T.; El-Haou, S.; Jackson, C.; Gkatzis, K.; Elliott, D.A.; Chuva de Sousa Lopes, S.M.; Mummery, C.L.; et al. Atrial-like cardiomyocytes from human pluripotent stem cells are a robust preclinical model for assessing atrial-selective pharmacology. EMBO Mol. Med. 2015, 7, 394–410. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Jiang, J.; Han, P.; Yuan, Q.; Zhang, J.; Zhang, X.; Xu, Y.; Cao, H.; Meng, Q.; Chen, L.; et al. Direct differentiation of atrial and ventricular myocytes from human embryonic stem cells by alternating retinoid signals. Cell Res. 2011, 21, 579–587. [Google Scholar] [CrossRef] [Green Version]
- Karakikes, I.; Senyei, G.D.; Hansen, J.; Kong, C.W.; Azeloglu, E.U.; Stillitano, F.; Lieu, D.K.; Wang, J.; Ren, L.; Hulot, J.S.; et al. Small molecule-mediated directed differentiation of human embryonic stem cells toward ventricular cardiomyocytes. Stem Cells Transl. Med. 2014, 3, 18–31. [Google Scholar] [CrossRef]
- Beltrami, A.P.; Barlucchi, L.; Torella, D.; Baker, M.; Limana, F.; Chimenti, S.; Kasahara, H.; Rota, M.; Musso, E.; Urbanek, K.; et al. Adult cardiac stem cells are multipotent and support myocardial regeneration. Cell 2003, 114, 763–776. [Google Scholar] [CrossRef] [Green Version]
- Cianflone, E.; Aquila, I.; Scalise, M.; Marotta, P.; Torella, M.; Nadal-Ginard, B.; Torella, D. Molecular basis of functional myogenic specification of Bona Fide multipotent adult cardiac stem cells. Cell Cycle 2018, 17, 927–946. [Google Scholar] [CrossRef] [Green Version]
- Scalise, M.; Marino, F.; Cianflone, E.; Mancuso, T.; Marotta, P.; Aquila, I.; Torella, M.; Nadal-Ginard, B.; Torella, D. Heterogeneity of Adult Cardiac Stem Cells. Adv. Exp. Med. Biol. 2019, 1169, 141–178. [Google Scholar] [CrossRef] [PubMed]
- Scalise, M.; Torella, M.; Marino, F.; Ravo, M.; Giurato, G.; Vicinanza, C.; Cianflone, E.; Mancuso, T.; Aquila, I.; Salerno, L.; et al. Atrial myxomas arise from multipotent cardiac stem cells. Eur. Heart J. 2020, 41, 4332–4345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marotta, P.; Cianflone, E.; Aquila, I.; Vicinanza, C.; Scalise, M.; Marino, F.; Mancuso, T.; Torella, M.; Indolfi, C.; Torella, D. Combining cell and gene therapy to advance cardiac regeneration. Expert. Opin. Biol. Ther. 2018, 18, 409–423. [Google Scholar] [CrossRef]
- Mancuso, T.; Barone, A.; Salatino, A.; Molinaro, C.; Marino, F.; Scalise, M.; Torella, M.; De Angelis, A.; Urbanek, K.; Torella, D.; et al. Unravelling the Biology of Adult Cardiac Stem Cell-Derived Exosomes to Foster Endogenous Cardiac Regeneration and Repair. Int. J. Mol. Sci. 2020, 21, 3725. [Google Scholar] [CrossRef] [PubMed]
- Marino, F.; Scalise, M.; Cianflone, E.; Salerno, L.; Cappetta, D.; Salerno, N.; De Angelis, A.; Torella, D.; Urbanek, K. Physical Exercise and Cardiac Repair: The Potential Role of Nitric Oxide in Boosting Stem Cell Regenerative Biology. Antioxidants 2021, 10, 1002. [Google Scholar] [CrossRef] [PubMed]
- Vicinanza, C.; Aquila, I.; Cianflone, E.; Scalise, M.; Marino, F.; Mancuso, T.; Fumagalli, F.; Giovannone, E.D.; Cristiano, F.; Iaccino, E.; et al. Kit(cre) knock-in mice fail to fate-map cardiac stem cells. Nature 2018, 555, E1–E5. [Google Scholar] [CrossRef]
- Ellison, G.M.; Torella, D.; Karakikes, I.; Nadal-Ginard, B. Myocyte death and renewal: Modern concepts of cardiac cellular homeostasis. Nat. Clin. Pract. Cardiovasc. Med. 2007, 4 (Suppl. S1), S52–S59. [Google Scholar] [CrossRef] [PubMed]
- Leong, Y.Y.; Ng, W.H.; Ellison-Hughes, G.M.; Tan, J.J. Cardiac Stem Cells for Myocardial Regeneration: They Are Not Alone. Front. Cardiovasc. Med. 2017, 4, 47. [Google Scholar] [CrossRef] [Green Version]
- Chimenti, I.; Gaetani, R.; Barile, L.; Forte, E.; Ionta, V.; Angelini, F.; Frati, G.; Messina, E.; Giacomello, A. Isolation and expansion of adult cardiac stem/progenitor cells in the form of cardiospheres from human cardiac biopsies and murine hearts. Methods Mol. Biol. 2012, 879, 327–338. [Google Scholar] [CrossRef]
- Urbanek, K.; Cesselli, D.; Rota, M.; Nascimbene, A.; De Angelis, A.; Hosoda, T.; Bearzi, C.; Boni, A.; Bolli, R.; Kajstura, J.; et al. Stem cell niches in the adult mouse heart. Proc. Natl. Acad. Sci. USA 2006, 103, 9226–9231. [Google Scholar] [CrossRef] [Green Version]
- Li, T.S.; Cheng, K.; Lee, S.T.; Matsushita, S.; Davis, D.; Malliaras, K.; Zhang, Y.; Matsushita, N.; Smith, R.R.; Marban, E. Cardiospheres recapitulate a niche-like microenvironment rich in stemness and cell-matrix interactions, rationalizing their enhanced functional potency for myocardial repair. Stem Cells 2010, 28, 2088–2098. [Google Scholar] [CrossRef] [Green Version]
- Davis, D.R.; Zhang, Y.; Smith, R.R.; Cheng, K.; Terrovitis, J.; Malliaras, K.; Li, T.S.; White, A.; Makkar, R.; Marbán, E. Validation of the cardiosphere method to culture cardiac progenitor cells from myocardial tissue. PLoS ONE 2009, 4, e7195. [Google Scholar] [CrossRef] [Green Version]
- Scalise, M.; Marino, F.; Salerno, L.; Mancuso, T.; Cappetta, D.; Barone, A.; Parrotta, E.I.; Torella, A.; Palumbo, D.; Veltri, P.; et al. In vitro CSC-derived cardiomyocytes exhibit the typical microRNA-mRNA blueprint of endogenous cardiomyocytes. Commun. Biol. 2021, 4, 1146. [Google Scholar] [CrossRef]
- Smith, R.R.; Barile, L.; Cho, H.C.; Leppo, M.K.; Hare, J.M.; Messina, E.; Giacomello, A.; Abraham, M.R.; Marban, E. Regenerative potential of cardiosphere-derived cells expanded from percutaneous endomyocardial biopsy specimens. Circulation 2007, 115, 896–908. [Google Scholar] [CrossRef] [Green Version]
- Chimenti, I.; Smith, R.R.; Li, T.S.; Gerstenblith, G.; Messina, E.; Giacomello, A.; Marban, E. Relative roles of direct regeneration versus paracrine effects of human cardiosphere-derived cells transplanted into infarcted mice. Circ. Res. 2010, 106, 971–980. [Google Scholar] [CrossRef]
- Cheng, K.; Li, T.S.; Malliaras, K.; Davis, D.R.; Zhang, Y.; Marban, E. Magnetic targeting enhances engraftment and functional benefit of iron-labeled cardiosphere-derived cells in myocardial infarction. Circ. Res. 2010, 106, 1570–1581. [Google Scholar] [CrossRef] [Green Version]
- Davis, D.R.; Kizana, E.; Terrovitis, J.; Barth, A.S.; Zhang, Y.; Smith, R.R.; Miake, J.; Marban, E. Isolation and expansion of functionally-competent cardiac progenitor cells directly from heart biopsies. J. Mol. Cell Cardiol. 2010, 49, 312–321. [Google Scholar] [CrossRef] [Green Version]
- Terrovitis, J.; Lautamaki, R.; Bonios, M.; Fox, J.; Engles, J.M.; Yu, J.; Leppo, M.K.; Pomper, M.G.; Wahl, R.L.; Seidel, J.; et al. Noninvasive quantification and optimization of acute cell retention by in vivo positron emission tomography after intramyocardial cardiac-derived stem cell delivery. J. Am. Coll. Cardiol. 2009, 54, 1619–1626. [Google Scholar] [CrossRef] [Green Version]
- Johnston, P.V.; Sasano, T.; Mills, K.; Evers, R.; Lee, S.T.; Smith, R.R.; Lardo, A.C.; Lai, S.; Steenbergen, C.; Gerstenblith, G.; et al. Engraftment, differentiation, and functional benefits of autologous cardiosphere-derived cells in porcine ischemic cardiomyopathy. Circulation 2009, 120, 1075–1083. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.T.; White, A.J.; Matsushita, S.; Malliaras, K.; Steenbergen, C.; Zhang, Y.; Li, T.S.; Terrovitis, J.; Yee, K.; Simsir, S.; et al. Intramyocardial injection of autologous cardiospheres or cardiosphere-derived cells preserves function and minimizes adverse ventricular remodeling in pigs with heart failure post-myocardial infarction. J. Am. Coll. Cardiol. 2011, 57, 455–465. [Google Scholar] [CrossRef] [Green Version]
- Takehara, N.; Tsutsumi, Y.; Tateishi, K.; Ogata, T.; Tanaka, H.; Ueyama, T.; Takahashi, T.; Takamatsu, T.; Fukushima, M.; Komeda, M.; et al. Controlled delivery of basic fibroblast growth factor promotes human cardiosphere-derived cell engraftment to enhance cardiac repair for chronic myocardial infarction. J. Am. Coll. Cardiol. 2008, 52, 1858–1865. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, X.; Pabon, L.; Murry, C.E. Engineering adolescence: Maturation of human pluripotent stem cell-derived cardiomyocytes. Circ. Res. 2014, 114, 511–523. [Google Scholar] [CrossRef] [Green Version]
- Mills, R.J.; Voges, H.K.; Porrello, E.R.; Hudson, J.E. Disease modeling and functional screening using engineered heart tissue. Curr. Opin. Physiol. 2018, 1, 80–88. [Google Scholar] [CrossRef]
- Horvath, P.; Aulner, N.; Bickle, M.; Davies, A.M.; Nery, E.D.; Ebner, D.; Montoya, M.C.; Ostling, P.; Pietiainen, V.; Price, L.S.; et al. Screening out irrelevant cell-based models of disease. Nat. Rev. Drug Discov. 2016, 15, 751–769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jabs, J.; Zickgraf, F.M.; Park, J.; Wagner, S.; Jiang, X.; Jechow, K.; Kleinheinz, K.; Toprak, U.H.; Schneider, M.A.; Meister, M.; et al. Screening drug effects in patient-derived cancer cells links organoid responses to genome alterations. Mol. Syst. Biol. 2017, 13, 955. [Google Scholar] [CrossRef] [PubMed]
- Moffat, J.G.; Vincent, F.; Lee, J.A.; Eder, J.; Prunotto, M. Opportunities and challenges in phenotypic drug discovery: An industry perspective. Nat. Rev. Drug Discov. 2017, 16, 531–543. [Google Scholar] [CrossRef]
- Zhao, D.; Lei, W.; Hu, S. Cardiac organoid—A promising perspective of preclinical model. Stem Cell Res. Ther. 2021, 12, 272. [Google Scholar] [CrossRef]
- Richards, D.J.; Li, Y.; Kerr, C.M.; Yao, J.; Beeson, G.C.; Coyle, R.C.; Chen, X.; Jia, J.; Damon, B.; Wilson, R.; et al. Human cardiac organoids for the modelling of myocardial infarction and drug cardiotoxicity. Nat. Biomed. Eng. 2020, 4, 446–462. [Google Scholar] [CrossRef]
- Shkumatov, A.; Baek, K.; Kong, H. Matrix rigidity-modulated cardiovascular organoid formation from embryoid bodies. PLoS ONE 2014, 9, e94764. [Google Scholar] [CrossRef]
- Tiburcy, M.; Meyer, T.; Soong, P.L.; Zimmermann, W.H. Collagen-based engineered heart muscle. Methods Mol. Biol. 2014, 1181, 167–176. [Google Scholar] [CrossRef]
- Ronaldson-Bouchard, K.; Vunjak-Novakovic, G. Organs-on-a-Chip: A Fast Track for Engineered Human Tissues in Drug Development. Cell Stem Cell 2018, 22, 310–324. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Rafatian, N.; Feric, N.T.; Cox, B.J.; Aschar-Sobbi, R.; Wang, E.Y.; Aggarwal, P.; Zhang, B.; Conant, G.; Ronaldson-Bouchard, K.; et al. A Platform for Generation of Chamber-Specific Cardiac Tissues and Disease Modeling. Cell 2019, 176, 913–927.e18. [Google Scholar] [CrossRef] [Green Version]
- Giacomelli, E.; Bellin, M.; Sala, L.; van Meer, B.J.; Tertoolen, L.G.; Orlova, V.V.; Mummery, C.L. Three-dimensional cardiac microtissues composed of cardiomyocytes and endothelial cells co-differentiated from human pluripotent stem cells. Development 2017, 144, 1008–1017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giacomelli, E.; Meraviglia, V.; Campostrini, G.; Cochrane, A.; Cao, X.; van Helden, R.W.J.; Krotenberg Garcia, A.; Mircea, M.; Kostidis, S.; Davis, R.P.; et al. Human-iPSC-Derived Cardiac Stromal Cells Enhance Maturation in 3D Cardiac Microtissues and Reveal Non-cardiomyocyte Contributions to Heart Disease. Cell Stem Cell 2020, 26, 862–879.e11. [Google Scholar] [CrossRef]
- Drakhlis, L.; Biswanath, S.; Farr, C.M.; Lupanow, V.; Teske, J.; Ritzenhoff, K.; Franke, A.; Manstein, F.; Bolesani, E.; Kempf, H.; et al. Human heart-forming organoids recapitulate early heart and foregut development. Nat. Biotechnol. 2021, 39, 737–746. [Google Scholar] [CrossRef] [PubMed]
- Rossi, G.; Broguiere, N.; Miyamoto, M.; Boni, A.; Guiet, R.; Girgin, M.; Kelly, R.G.; Kwon, C.; Lutolf, M.P. Capturing Cardiogenesis in Gastruloids. Cell Stem Cell 2021, 28, 230–240.e6. [Google Scholar] [CrossRef]
- Meilhac, S.M.; Buckingham, M.E. The deployment of cell lineages that form the mammalian heart. Nat. Rev. Cardiol. 2018, 15, 705–724. [Google Scholar] [CrossRef]
- Lewis-Israeli, Y.R.; Volmert, B.D.; Gabalski, M.A.; Huang, A.R.; Aguirre, A. Generating Self-Assembling Human Heart Organoids Derived from Pluripotent Stem Cells. J. Vis. Exp. 2021. [Google Scholar] [CrossRef]
- Hoang, P.; Kowalczewski, A.; Sun, S.; Winston, T.S.; Archilla, A.M.; Lemus, S.M.; Ercan-Sencicek, A.G.; Gupta, A.R.; Liu, W.; Kontaridis, M.I.; et al. Engineering spatial-organized cardiac organoids for developmental toxicity testing. Stem Cell Rep. 2021, 16, 1228–1244. [Google Scholar] [CrossRef] [PubMed]
- Mills, R.J.; Titmarsh, D.M.; Koenig, X.; Parker, B.L.; Ryall, J.G.; Quaife-Ryan, G.A.; Voges, H.K.; Hodson, M.P.; Ferguson, C.; Drowley, L.; et al. Functional screening in human cardiac organoids reveals a metabolic mechanism for cardiomyocyte cell cycle arrest. Proc. Natl. Acad. Sci. USA 2017, 114, E8372–E8381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Voges, H.K.; Mills, R.J.; Elliott, D.A.; Parton, R.G.; Porrello, E.R.; Hudson, J.E. Development of a human cardiac organoid injury model reveals innate regenerative potential. Development 2017, 144, 1118–1127. [Google Scholar] [CrossRef] [Green Version]
- Nugraha, B.; Buono, M.F.; Emmert, M.Y. Modelling human cardiac diseases with 3D organoid. Eur. Heart J. 2018, 39, 4234–4237. [Google Scholar] [CrossRef]
- Mills, R.J.; Parker, B.L.; Quaife-Ryan, G.A.; Voges, H.K.; Needham, E.J.; Bornot, A.; Ding, M.; Andersson, H.; Polla, M.; Elliott, D.A.; et al. Drug Screening in Human PSC-Cardiac Organoids Identifies Pro-proliferative Compounds Acting via the Mevalonate Pathway. Cell Stem Cell 2019, 24, 895–907.e6. [Google Scholar] [CrossRef] [PubMed]
- Nugraha, B.; Buono, M.F.; von Boehmer, L.; Hoerstrup, S.P.; Emmert, M.Y. Human Cardiac Organoids for Disease Modeling. Clin. Pharmacol. Ther. 2019, 105, 79–85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Filippo Buono, M.; von Boehmer, L.; Strang, J.; Hoerstrup, S.P.; Emmert, M.Y.; Nugraha, B. Human Cardiac Organoids for Modeling Genetic Cardiomyopathy. Cells 2020, 9, 1733. [Google Scholar] [CrossRef]
- Forte, E.; Furtado, M.B.; Rosenthal, N. The interstitium in cardiac repair: Role of the immune-stromal cell interplay. Nat. Rev. Cardiol. 2018, 15, 601–616. [Google Scholar] [CrossRef]
- Passier, R.; Orlova, V.; Mummery, C. Complex Tissue and Disease Modeling using hiPSCs. Cell Stem Cell 2016, 18, 309–321. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Sutani, A.; Kaneko, R.; Takeuchi, J.; Sasano, T.; Kohda, T.; Ihara, K.; Takahashi, K.; Yamazoe, M.; Morio, T.; et al. In vitro generation of functional murine heart organoids via FGF4 and extracellular matrix. Nat. Commun. 2020, 11, 4283. [Google Scholar] [CrossRef] [PubMed]
- Long, C.; Li, H.; Tiburcy, M.; Rodriguez-Caycedo, C.; Kyrychenko, V.; Zhou, H.; Zhang, Y.; Min, Y.L.; Shelton, J.M.; Mammen, P.P.A.; et al. Correction of diverse muscular dystrophy mutations in human engineered heart muscle by single-site genome editing. Sci. Adv. 2018, 4, eaap9004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shinnawi, R.; Shaheen, N.; Huber, I.; Shiti, A.; Arbel, G.; Gepstein, A.; Ballan, N.; Setter, N.; Tijsen, A.J.; Borggrefe, M.; et al. Modeling Reentry in the Short QT Syndrome With Human-Induced Pluripotent Stem Cell-Derived Cardiac Cell Sheets. J. Am. Coll. Cardiol. 2019, 73, 2310–2324. [Google Scholar] [CrossRef]
- Wang, G.; McCain, M.L.; Yang, L.; He, A.; Pasqualini, F.S.; Agarwal, A.; Yuan, H.; Jiang, D.; Zhang, D.; Zangi, L.; et al. Modeling the mitochondrial cardiomyopathy of Barth syndrome with induced pluripotent stem cell and heart-on-chip technologies. Nat. Med. 2014, 20, 616–623. [Google Scholar] [CrossRef] [PubMed]
- Eder, A.; Vollert, I.; Hansen, A.; Eschenhagen, T. Human engineered heart tissue as a model system for drug testing. Adv. Drug. Deliv. Rev. 2016, 96, 214–224. [Google Scholar] [CrossRef]
- Marino, F.; Scalise, M.; Cianflone, E.; Mancuso, T.; Aquila, I.; Agosti, V.; Torella, M.; Paolino, D.; Mollace, V.; Nadal-Ginard, B.; et al. Role of c-Kit in Myocardial Regeneration and Aging. Front. Endocrinol. (Lausanne) 2019, 10, 371. [Google Scholar] [CrossRef]
- Nadal-Ginard, B.; Cianflone, E.; Torella, D. The baby and the bath water: Adult cardiac stem cells revisited. Eur. Heart J. 2021, 42, 3814–3816. [Google Scholar] [CrossRef]
- Cianflone, E.; Scalise, M.; Salerno, N.; Torella, D. Cardiac stem cell therapy towards the clinic: The way forward re-starts from within. Int. J. Cardiol. 2021. [Google Scholar] [CrossRef] [PubMed]
- Thijssen, D.H.; Torella, D.; Hopman, M.T.; Ellison, G.M. The role of endothelial progenitor and cardiac stem cells in the cardiovascular adaptations to age and exercise. Front. Biosci. (Landmark Ed.) 2009, 14, 4685–4702. [Google Scholar] [CrossRef] [Green Version]
- Cianflone, E.; Torella, M.; Chimenti, C.; De Angelis, A.; Beltrami, A.P.; Urbanek, K.; Rota, M.; Torella, D. Adult Cardiac Stem Cell Aging: A Reversible Stochastic Phenomenon? Oxid. Med. Cell. Longev. 2019, 2019, 5813147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aquila, I.; Cianflone, E.; Scalise, M.; Marino, F.; Mancuso, T.; Filardo, A.; Smith, A.J.; Cappetta, D.; De Angelis, A.; Urbanek, K.; et al. c-kit Haploinsufficiency impairs adult cardiac stem cell growth, myogenicity and myocardial regeneration. Cell Death Dis. 2019, 10, 436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cianflone, E.; Cappetta, D.; Mancuso, T.; Sabatino, J.; Marino, F.; Scalise, M.; Albanese, M.; Salatino, A.; Parrotta, E.I.; Cuda, G.; et al. Statins Stimulate New Myocyte Formation After Myocardial Infarction by Activating Growth and Differentiation of the Endogenous Cardiac Stem Cells. Int. J. Mol. Sci. 2020, 21, 7927. [Google Scholar] [CrossRef]
- Cianflone, E.; Torella, M.; Biamonte, F.; De Angelis, A.; Urbanek, K.; Costanzo, F.S.; Rota, M.; Ellison-Hughes, G.M.; Torella, D. Targeting Cardiac Stem Cell Senescence to Treat Cardiac Aging and Disease. Cells 2020, 9, 1558. [Google Scholar] [CrossRef] [PubMed]
- Torella, D.; Iaconetti, C.; Tarallo, R.; Marino, F.; Giurato, G.; Veneziano, C.; Aquila, I.; Scalise, M.; Mancuso, T.; Cianflone, E.; et al. miRNA Regulation of the Hyperproliferative Phenotype of Vascular Smooth Muscle Cells in Diabetes. Diabetes 2018, 67, 2554–2568. [Google Scholar] [CrossRef] [Green Version]
- Stevens, K.R.; Pabon, L.; Muskheli, V.; Murry, C.E. Scaffold-free human cardiac tissue patch created from embryonic stem cells. Tissue Eng. Part. A 2009, 15, 1211–1222. [Google Scholar] [CrossRef]
- Yui, S.; Nakamura, T.; Sato, T.; Nemoto, Y.; Mizutani, T.; Zheng, X.; Ichinose, S.; Nagaishi, T.; Okamoto, R.; Tsuchiya, K.; et al. Functional engraftment of colon epithelium expanded in vitro from a single adult Lgr5(+) stem cell. Nat. Med. 2012, 18, 618–623. [Google Scholar] [CrossRef]
- Fowler, J.L.; Ang, L.T.; Loh, K.M. A critical look: Challenges in differentiating human pluripotent stem cells into desired cell types and organoids. Wiley Interdiscip. Rev. Dev. Biol. 2020, 9, e368. [Google Scholar] [CrossRef] [PubMed]
- Lewis-Israeli, Y.R.; Wasserman, A.H.; Aguirre, A. Heart Organoids and Engineered Heart Tissues: Novel Tools for Modeling Human Cardiac Biology and Disease. Biomolecules 2021, 11, 1277. [Google Scholar] [CrossRef] [PubMed]
- Tulloch, N.L.; Muskheli, V.; Razumova, M.V.; Korte, F.S.; Regnier, M.; Hauch, K.D.; Pabon, L.; Reinecke, H.; Murry, C.E. Growth of engineered human myocardium with mechanical loading and vascular coculture. Circ. Res. 2011, 109, 47–59. [Google Scholar] [CrossRef] [PubMed]
- Yin, F.; Zhang, X.; Wang, L.; Wang, Y.; Zhu, Y.; Li, Z.; Tao, T.; Chen, W.; Yu, H.; Qin, J. HiPSC-derived multi-organoids-on-chip system for safety assessment of antidepressant drugs. Lab Chip 2021, 21, 571–581. [Google Scholar] [CrossRef]
- Miranda, C.C.; Fernandes, T.G.; Diogo, M.M.; Cabral, J.M.S. Towards Multi-Organoid Systems for Drug Screening Applications. Bioengineering 2018, 5, 49. [Google Scholar] [CrossRef] [Green Version]



| Methodology | Growth Characteristics | Advantages | Disvantages |
|---|---|---|---|
| Suspension cultures | Uncontrolled spheroid size, non-uniform spheroid shape, variations in cell number | Simple, cheap, quick to scale up | Low efficiency |
| Hanging drops | Fast spheroid formation, well-controlled and uniform spheroid size | Simple, no special equipment is required | Laborious task, time-consuming, instable, difficult long-term cultures, low throughput, low efficiency |
| Forced aggregation | Rapid cell aggregation, well-controlled and uniform spheroid size | Increased efficiency, high throughput | More complex, undefined potential effects to cells, special equipment and culture conditions are required |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Scalise, M.; Marino, F.; Salerno, L.; Cianflone, E.; Molinaro, C.; Salerno, N.; De Angelis, A.; Viglietto, G.; Urbanek, K.; Torella, D. From Spheroids to Organoids: The Next Generation of Model Systems of Human Cardiac Regeneration in a Dish. Int. J. Mol. Sci. 2021, 22, 13180. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms222413180
Scalise M, Marino F, Salerno L, Cianflone E, Molinaro C, Salerno N, De Angelis A, Viglietto G, Urbanek K, Torella D. From Spheroids to Organoids: The Next Generation of Model Systems of Human Cardiac Regeneration in a Dish. International Journal of Molecular Sciences. 2021; 22(24):13180. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms222413180
Chicago/Turabian StyleScalise, Mariangela, Fabiola Marino, Luca Salerno, Eleonora Cianflone, Claudia Molinaro, Nadia Salerno, Antonella De Angelis, Giuseppe Viglietto, Konrad Urbanek, and Daniele Torella. 2021. "From Spheroids to Organoids: The Next Generation of Model Systems of Human Cardiac Regeneration in a Dish" International Journal of Molecular Sciences 22, no. 24: 13180. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms222413180