Advances in Plant Metabolomics and Its Applications in Stress and Single-Cell Biology
Abstract
:1. Introduction
2. Metabolomics Technologies and Advancements
3. Metabolomics Applications in Plant Stress Responses, Multi-Omics and Single-Cell Biology
3.1. Diversity of Plant Metabolites as a Result of Ecological Adaptation
3.2. Dynamics of Plant Metabolites in Response to Stresses
3.2.1. Abiotic Stresses
Drought, Flooding and Heat Stress
Salinity Stress
Metal, Atmospheric, and Nutrient Stresses
3.2.2. Biotic Stress Resistance in Plants
Metabolomics of Plant-Microbe Interactions
Metabolomics of Plant-Herbivore Interactions
Metabolomics of Hormonal Crosstalk
3.3. Plant Interactions with Multiple Abiotic and Biotic Stresses
3.4. Metabolomics of Chemical Agent Treatment and Epigenetic Modifications
3.5. Multi-Omics Integration to Analyze Plant Multiple Stress Responses
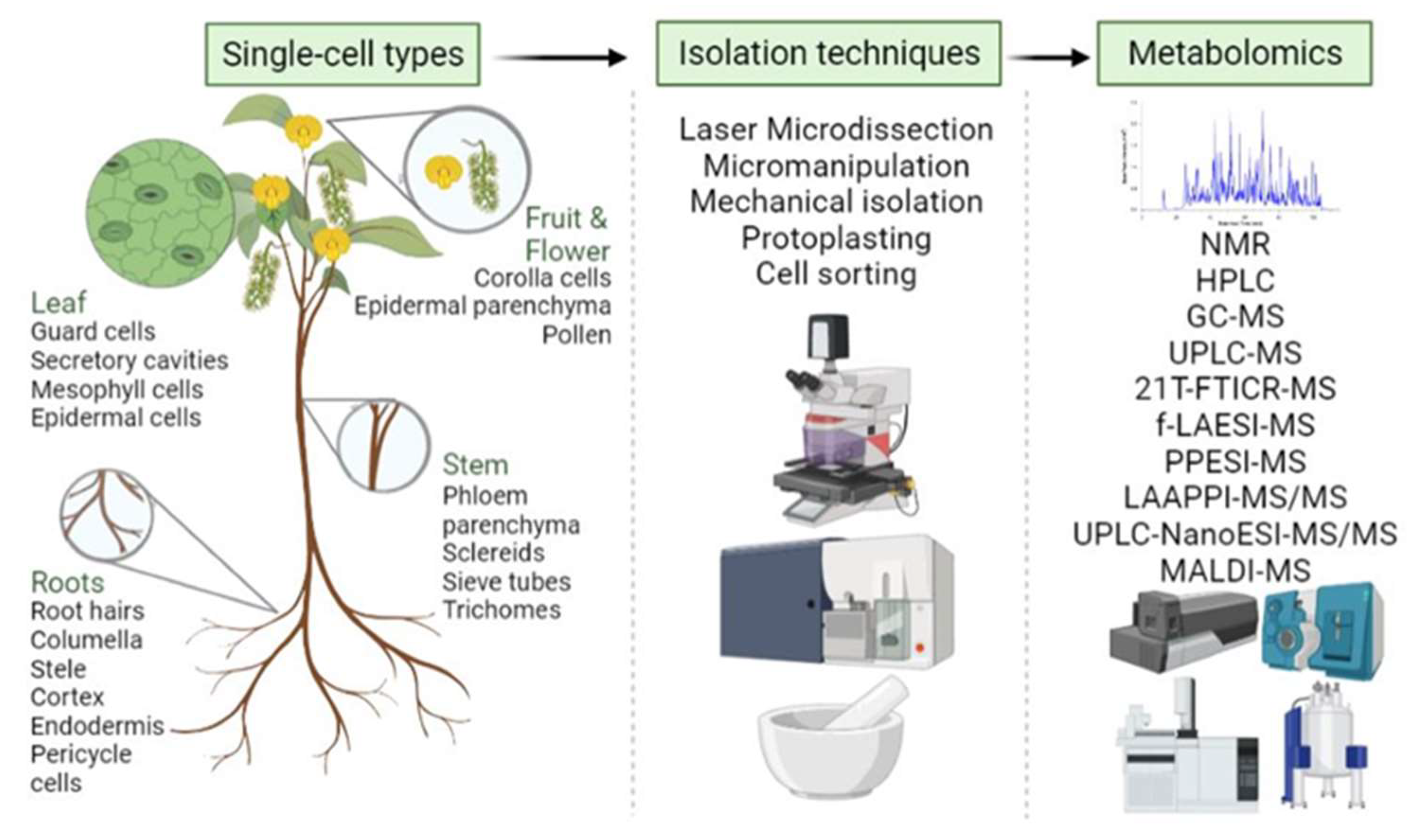
3.6. Single-Cell and Spatial Metabolomics
4. Challenges and Future Perspectives
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Delepine, M.J. Josehp Pelletier and Joseph Caventou. Chem. Educ. 1951, 28, 454–461. [Google Scholar] [CrossRef]
- Misra, B.B.; Chen, S. Advances in understanding CO2 responsive plant metabolomes in the era of climate change. Metabolomics 2015, 11, 1478–1491. [Google Scholar] [CrossRef]
- Sumner, L.W.; Lei, Z.; Nikolau, B.J.; Saito, K. Modern plant metabolomics: Advanced natural product gene discoveries, improved technologies, and future prospects. Nat. Prod. Rep. 2015, 32, 212–229. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Han, H.; Liu, A.; Guan, Q.; Kang, J.; David, L.; Dufresne, C.; Chen, S.; Tian, J. Combined ultraviolet and darkness regulation of medicinal metabolites in Mehonia bealei revealed by proteomics and metabolomics. J. Proteom. 2021, 233, 104081. [Google Scholar] [CrossRef]
- Holland, J.F.; Leary, J.J.; Sweeley, C.C. Advanced instrumentation and strategies for metabolic profiling. J. Chromatogr. B Biomed. Sci. Appl. 1986, 379, 3–26. [Google Scholar] [CrossRef]
- Sauter, H.; Lauer, M.; Fritsch, H. Metabolic Profiling of Plants: A New Diagnostic Technique; ACS Symposium Series-American Chemical Society (USA): Washington, DC, USA, 1991; Volume 2, pp. 49–65. [Google Scholar]
- Fiehn, O.; Kopka, J.; Dörmann, P.; Altmann, T.; Trethewey, R.N.; Willmitzer, L. Metabolite profiling for plant functional genomics. Nat. Biotechnol. 2000, 18, 1157–1161. [Google Scholar] [CrossRef]
- Roessner, U.; Wagner, C.; Kopka, J.; Trethewey, R.N.; Willmitzer, L. Simultaneous analysis of metabolites in potato tuber by gas chromatography-mass spectrometry. Plant J. 2000, 23, 131–142. [Google Scholar] [CrossRef]
- Larson, T.R.; Graham, I.A. Technical advance: A novel technique for the sensitive quantification of acyl CoA esters from plant tissues. Plant J. 2001, 25, 115–125. [Google Scholar] [CrossRef]
- Hall, R.; Beale, M.; Fiehn, O.; Hardy, N.; Sumner, L.; Bino, R. Plant metabolomics: The missing link in functional genomics strategies. Plant Cell 2002, 14, 1437–1440. [Google Scholar] [CrossRef] [Green Version]
- Bailey, N.J.; Oven, M.; Holmes, E.; Nicholson, J.K.; Zenk, M.H. Metabolomic analysis of the consequences of cadmium exposure in Silene cucubalus cell cultures via 1H NMR spectroscopy and chemometrics. Phytochemistry 2003, 62, 851–858. [Google Scholar] [CrossRef]
- Defernez, M.; Colquhoun, I.J. Factors affecting the robustness of metabolite fingerprinting using 1H NMR spectra. Phytochemistry 2003, 62, 1009–1017. [Google Scholar] [CrossRef]
- Schroeder, M.; Meyer, S.W.; Heyman, H.M.; Barsch, A.; Sumner, L.W. Generation of a collision cross-section library for multi-dimensional plant metabolomics using UHPLC-trapped ion mobility-MS/MS. Metabolites 2020, 10, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guijas, C.; Montenegro-Burke, J.R.; Warth, B.; Spilker, M.E.; Siuzdak, G. Metabolomics activity screening for identifying metabolites that modulate phenotype. Nat. Biotechnol. 2018, 36, 316–320. [Google Scholar] [CrossRef] [PubMed]
- Alseekh, S.; Fernie, A.R. Metabolomics 20 years on: What have we learned and what hurdles remain? Plant J. 2018, 94, 933–942. [Google Scholar] [CrossRef]
- Geng, S.; Misra, B.B.; de Armas, E.; Huhman, D.V.; Alborn, H.T.; Sumner, L.W.; Chen, S. Jasmonate-mediated stomatal closure under elevated CO2 revealed by time-resolved metabolomics. Plant J. 2016, 88, 947–962. [Google Scholar] [CrossRef] [Green Version]
- Geng, S.; Yu, B.; Zhu, N.; Dufresne, C.; Chen, S. Metabolomics and proteomics of Brassica napus guard cells in response to low CO2. Front. Mol. Biosci. 2017, 4, 51. [Google Scholar] [CrossRef] [Green Version]
- Zhu, M.; Geng, S.; Chakravorty, D.; Guan, Q.; Chen, S.; Assmann, S.M. Metabolomics of red-light-induced stomatal opening in Arabidopsis thaliana: Coupling with abscisic acid and jasmonic acid metabolism. Plant J. 2019, 101, 1331–1348. [Google Scholar] [CrossRef]
- Mwendwa, M.; Weston, P.A.; Fomsgaard, I.; Laursen, B.B.; Brown, W.B.; Wu, H.; Rebetzke, G.; Jane, C.; Quinn, J.C.Q.; Weston, L.A. Metabolic profiling for benzoxazinoids in weed-suppressive and early vigour wheat genotypes. In Proceedings of the Twentieth Australasian Weeds Conference, Perth, WA, Australia, 11–15 September 2018; pp. 353–357. [Google Scholar]
- Glover, S.C.; Nouri, M.Z.; Tuna, K.M.; Mendoza Alvarez, L.B.; Ryan, L.K.; Shirley, J.F.; Tang, Y.; Denslow, N.D.; Alli, A.A. Lipidomic analysis of urinary exosomes from hereditary α-tryptasemia patients and healthy volunteers. FASEB BioAdvances 2019, 10, 624–638. [Google Scholar] [CrossRef]
- Dreier, D.A.; Nouri, M.Z.; Denslow, N.D.; Martyniuk, C.J. Lipidomics reveals multiple stressor effects (temperature × mitochondrial toxicant) in the zebrafish embryo toxicity test. Chemosphere 2021, 264, 128472. [Google Scholar] [CrossRef]
- Billet, K.; Malinowska, M.A.; Munsch, T.; Unlubayir, M.; Adler, S.; Delanoue, G.; LaNoue, A. Semi-Targeted Metabolomics to Validate Biomarkers of Grape Downy Mildew Infection Under Field Conditions. Plants 2020, 9, 1008. [Google Scholar] [CrossRef]
- Zheng, F.; Zhao, X.; Zeng, Z.; Wang, L.; Lv, W.; Wang, Q.; Xu, G. Development of a plasma pseudo targeted metabolomics method based on ultra-high-performance liquid chromatography–mass spectrometry. Nat. Protoc. 2020, 15, 2519–2537. [Google Scholar] [CrossRef] [PubMed]
- Gowda, G.A.N.; Djukovic, D. Overview of mass spectrometry-based metabolomics: Opportunities and challenges. Methods Mol. Biol. 2014, 1198, 3–12. [Google Scholar] [PubMed] [Green Version]
- Stettin, D.; Poulin, R.X.; Pohnert, G. Metabolomics benefits from Orbitrap GC–MS—comparison of low- and high-resolution GC–MS. Metabolites 2020, 10, 143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeki, Ö.C.; Eylem, C.C.; Reçber, T.; Kır, S.; Nemutlu, E. Integration of GC–MS and LC–MS for untargeted metabolomics profiling. J. Pharm. Biomed. Anal. 2020, 190, 113509. [Google Scholar] [CrossRef] [PubMed]
- Tabassam, Q.; Mehmood, T.; Ahmed, S.; Saeed, S.; Raza, A.R.; Anwar, F. GC-MS metabolomics profiling and HR-APCI-MS characterization of potential anticancer compounds and antimicrobial activities of extracts from Picrorhiza kurroa roots. J. Appl. Biomed. 2021, 19, 26–39. [Google Scholar] [CrossRef] [PubMed]
- Fagbohun, O.F.; Olawoye, B.; Ademakinwa, A.N.; Oriyomi, O.V.; Fagbohun, O.S.; Fadare, O.A.; Msagati, T.A. UHPLC/GC-TOF-MS metabolomics, MTT assay, and molecular docking studies reveal physostigmine as a new anticancer agent from the ethyl acetate and butanol fractions of Kigelia africana (Lam.) Benth. fruit extracts. Biomed. Chromatogr. 2021, 35, 2. [Google Scholar] [CrossRef] [PubMed]
- Lima, V.F.; Erban, A.; Daubermann, A.G.; Freire, F.B.S.; Porto, N.P.; Cândido-Sobrinho, S.A.; Medeiros, D.B.; Schwarzländer, M.; Fernie, A.R.; Dos Anjos, L.; et al. Establishment of a GC-MS-based 13 C-positional isotopomer approach suitable for investigating metabolic fluxes in plant primary metabolism. Plant J. 2021, 50, 2000–2010. [Google Scholar] [CrossRef]
- Feizi, N.; Hashemi-Nasab, F.S.; Golpelichi, F.; Saburouh, N.; Parastar, H. Recent trends in application of chemometric methods for GC-MS and GC×GC-MS-based metabolomic studies. TrAC Trends Anal. Chem. 2021, 138, 116239. [Google Scholar] [CrossRef]
- Lim, V.; Gorji, S.G.; Daygon, V.D.; Fitzgerald, M. Untargeted and targeted metabolomic profiling of Australian indigenous fruits. Metabolites 2020, 10, 114. [Google Scholar] [CrossRef] [Green Version]
- Motshudi, M.C.; Olaokun, O.O.; Mkolo, N.M. Evaluation of GC × GC-TOF-MS untargeted metabolomics, cytotoxicity and antimicrobial activity of leaf extracts of Artemisia afra (Jacq.) purchased from three local vendors. J. King Saud Univ.-Sci. 2021, 33, 101422. [Google Scholar] [CrossRef]
- Misra, B.B.; Bassey, E.; Bishop, A.C.; Kusel, D.T.; Cox, L.A.; Olivier, M. High-resolution gas chromatography/mass spectrometry metabolomics of non-human primate serum. Rapid Commun. Mass Spectrom. 2018, 32, 1497–1506. [Google Scholar] [CrossRef] [PubMed]
- Peterson, A.C.; Hauschild, J.-P.; Quarmby, S.T.; Krumwiede, D.; Lange, O.; Lemke, R.A.S.; Grosse-Coosmann, F.; Horning, S.; Donohue, T.J.; Westphall, M.S.; et al. Development of a GC/Quadrupole-Orbitrap Mass Spectrometer, Part I: Design and Characterization. Anal. Chem. 2014, 86, 10036–10043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weidt, S.; Haggarty, J.; Kean, R.; Cojocariu, C.I.; Silcock, P.J.; Rajendran, R.; Ramage, G.; Burgess, K.E.V. A novel targeted/untargeted GC-Orbitrap metabolomics methodology applied to Candida albicans and Staphylococcus aureus biofilms. Metabolomics 2016, 12, 189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brockbals, L.; Habicht, M.; Hajdas, I.; Galassi, F.M.; Rühli, F.J.; Kraemer, T. Untargeted metabolomics-like screening approach for chemical characterization and differentiation of canopic jar and mummy samples from ancient Egypt using GC-high resolution MS. Analyst 2018, 143, 4503–4512. [Google Scholar] [CrossRef] [PubMed]
- Misra, B.B.; Olivier, M. High resolution GC-Orbitrap-MS metabolomics using both electron ionization and chemical ionization for analysis of human plasma. J. Proteome Res. 2020, 19, 2717–2731. [Google Scholar] [CrossRef] [PubMed]
- Misra, B.B. Advances in high resolution GC-MS technology: A focus on the application of GC-Orbitrap-MS in metabolomics and exposomics for FAIR practices. Anal. Methods 2021, 13, 2265–2282. [Google Scholar] [CrossRef]
- Romera-Torres, A.; Arrebola-Liébanas, J.; Vidal, J.L.M.; Frenich, A.G. Determination of calystegines in several tomato varieties based on GC-Q-Orbitrap analysis and their classification by ANOVA. J. Agric. Food Chem. 2019, 67, 1284–1291. [Google Scholar] [CrossRef]
- Rivera-Pérez, A.; Romero-González, R.; Garrido Frenich, A. Feasibility of applying untargeted metabolomics with GC-Orbitrap-HRMS and chemometrics for authentication of black pepper (Piper nigrum L.) and identification of geographical and processing markers. J. Agric. Food Chem. 2021, 69, 5547–5558. [Google Scholar] [CrossRef]
- Perez de Souza, L.; Alseekh, S.; Scossa, F.; Fernie, A.R. Ultra-high-performance liquid chromatography high-resolution mass spectrometry variants for metabolomics research. Nat. Methods 2021, 18, 733–746. [Google Scholar] [CrossRef]
- Wickremsinhe, E.; Singh, G.; Ackermann, B.; Gillespie, T.; Chaudhary, A. A review of nanoelectrospray ionization applications for drug metabolism and pharmacokinetics. Curr. Drug Metab. 2006, 7, 913–928. [Google Scholar] [CrossRef]
- Hilhorst, M.; Briscoe, C.; van de Merbel, N. Sense and nonsense of miniaturized LC–MS/MS for bioanalysis. Bioanalysis 2014, 6, 3263–3265. [Google Scholar] [CrossRef] [PubMed]
- Marginean, I.; Tang, K.; Smith, R.D.; Kelly, R.T. Picoelectrospray ionization mass spectrometry using narrow-bore chemically etched emitters. J. Am. Soc. Mass Spectrom. 2014, 25, 30–36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chetwynd, A.J.; David, A. A review of nanoscale LC-ESI for metabolomics and its potential to enhance the metabolome coverage. Talanta 2018, 182, 380–390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ringbeck, B.; Bury, D.; Hayen, H.; Weiss, T.; Brüning, T.; Koch, H.M. Determination of specific urinary nonylphenol metabolites by online-SPE-LC-MS/MS as novel human exposure biomarkers. J. Chromatogr. B 2021, 1177, 122794. [Google Scholar] [CrossRef]
- Deng, J.; Zhang, G.; Neubert, T.A. Metabolomic analysis of glioma cells using nanoflow liquid chromatography-tandem mass spectrometry. Methods Mol. Biol. 2018, 1741, 125–134. [Google Scholar]
- Žampachová, L.; Aturki, Z.; Mariani, F.; Bednář, P. A rapid nano-liquid chromatographic method for the analysis of cannabinoids in Cannabis sativa L. Extracts. Molecules 2021, 26, 1825. [Google Scholar] [CrossRef]
- Vasilopoulou, C.G.; Sulek, K.; Brunner, A.-D.; Meitei, N.S.; Schweiger-Hufnagel, U.; Meyer, S.W.; Barsch, A.; Mann, M.; Meier, F. Trapped ion mobility spectrometry and PASEF enable in-depth lipidomics from minimal sample amounts. Nat. Commun. 2020, 11, 331. [Google Scholar] [CrossRef] [Green Version]
- Delvaux, A.; Rathahao-Paris, E.; Alves, S. Different ion mobility-mass spectrometry coupling techniques to promote metabolomics. Mass Spectrom. Rev. 2021, 10, 1002. [Google Scholar] [CrossRef]
- Paglia, G.; Smith, A.J.; Astarita, G. Ion mobility mass spectrometry in the omics era: Challenges and opportunities for metabolomics and lipidomics. Mass Spectrom. Rev. 2021, 40, 1–44. [Google Scholar] [CrossRef]
- Falco, B.; Manzo, D.; Incerti, G.; Garonna, A.; Pietro Garonna, A.; Ercolano, M.; Lanzotti, V. Metabolomics approach based on NMR spectroscopy and multivariate data analysis to explore the interaction between the leafminer Tuta absoluta and tomato (Solanum lycopersicum). Phytochem. Anal. 2019, 30, 556–563. [Google Scholar] [CrossRef]
- Lacerda, J.W.F.; Siqueira, K.A.; Vasconcelos, L.G.; Bellete, B.S.; Dall’Oglio, E.L.; Sousa Junior, P.T.; Sampaio, O.M. Metabolomic analysis of combretum lanceolatum plants interaction with Diaporthe phaseolorum and Trichoderma spirale endophytic fungi through 1 H-NMR. Chem. Biodivers 2021, 18, e2100350. [Google Scholar] [CrossRef] [PubMed]
- Murti, R.H.; Afifah, E.N.; Nuringtyas, T.R. Metabolomic response of tomatoes (Solanum lycopersicum L.) against bacterial wilt (Ralstonia solanacearum) using 1H-NMR spectroscopy. Plants 2021, 10, 1143. [Google Scholar] [CrossRef] [PubMed]
- Bapela, M.J.; Heyman, H.; Senejoux, F.; Meyer, J.J.M. 1H NMR-based metabolomics of antimalarial plant species traditionally used by Vha-Venda people in Limpopo Province, South Africa and isolation of antiplasmodial compounds. J. Ethnopharmacol. 2019, 228, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Kim, S.-Y.; Lee, J.; Shin, B.; Seo, J.-A.; Kim, Y.-S.; Lee, D.; Choi, H.-K. Discrimination of the geographical origin of soybeans using NMR-based metabolomics. Foods 2021, 10, 435. [Google Scholar] [CrossRef] [PubMed]
- Knitsch, R.; AlWahsh, M.; Raschke, H.; Lambert, J.; Hergenröder, R. In vitro spatio-temporal NMR metabolomics of living 3D cell models. Anal. Chem. 2021, 93, 13485–13494. [Google Scholar] [CrossRef] [PubMed]
- Gebretsadik, T.; Linert, W.; Thomas, M.; Berhanu, T.; Frew, R. LC–NMR for natural product analysis: A journey from an academic curiosity to a robust analytical tool. Science 2021, 3, 6. [Google Scholar] [CrossRef]
- Marshall, D.D.; Powers, R. Beyond the paradigm: Combining mass spectrometry and nuclear. Physiol Behav 2017, 176, 139–148. [Google Scholar]
- Shi, B.; Ding, H.; Wang, L.; Wang, C.; Tian, X.; Fu, Z.; Zhang, L.; Han, L. Investigation on the stability in plant metabolomics with a special focus on freeze-thaw cycles: LC–MS and NMR analysis to Cassiae Semen (Cassia obtusifolia L.) seeds as a case study. J. Pharm. Biomed. Anal. 2021, 204, 114243. [Google Scholar] [CrossRef]
- Cody, R.B.; Laramée, J.A.; Durst, H.D. Versatile new ion source for the analysis of materials in open air under ambient conditions. Anal. Chem. 2005, 77, 2297–2302. [Google Scholar] [CrossRef]
- Novotna, H.; Kmiecik, O.; Galazka, M.; Krtkova, V.; Hurajova, A.; Schulzova, V.; Hallmann, E.; Rembialkowska, E.; Hajslova, J. Metabolomic fingerprinting employing DART-TOFMS for authentication of tomatoes and peppers from organic and conventional farming. Food Addit. Contam. Part A 2012, 29, 1335–1346. [Google Scholar] [CrossRef]
- Jehan, Z. Single-cell omics: An overview. Single Cell Omics 2019, 1, 3–19. [Google Scholar]
- Misra, B.B.; Assmann, S.M.; Chen, S. Plant single cell and single cell-type metabolomics. Trends Plant Sci. 2014, 19, 637–646. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, T.; Miyakawa, S.; Esaki, T.; Mizuno, H.; Masujima, T.; Koshiba, T.; Seo, M. Live single-cell plant hormone analysis by video-mass spectrometry. Plant Cell Physiol. 2015, 56, 1287–1296. [Google Scholar] [CrossRef] [Green Version]
- Fujii, T.; Matsuda, S.; Tejedor, M.L.; Esaki, T.; Sakane, I.; Mizuno, H.; Tsuyama, N.; Masujima, T. Direct metabolomics for plant cells by live single-cell mass spectrometry. Nat. Protoc. 2015, 10, 1445–1456. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, T.; Wada, H.; Morita, S.; Erra-Balsells, R.; Hiraoka, K.; Nonami, H. Single-cell metabolite profiling of stalk and glandular cells of intact trichomes with internal electrode capillary pressure probe electrospray ionization mass spectrometry. Anal. Chem. 2016, 88, 3049–3057. [Google Scholar] [CrossRef]
- Yamamoto, K.; Takahashi, K.; Caputi, L.; Mizuno, H.; Rodriguez-Lopez, C.E.; Iwasaki, T.; Mimura, T. The complexity of intercellular localization of alkaloids revealed by single-cell metabolomics. New Phytol. 2019, 224, 848–859. [Google Scholar] [CrossRef]
- Wada, H.; Hatakeyama, Y.; Onda, Y.; Nonami, H.; Nakashima, T.; Erra-Balsells, R.; Morita, S.; Hiraoka, K.; Tanaka, F.; Nakano, H. Multiple strategies for heat adaptation to prevent chalkiness in the rice endosperm. J. Exp. Bot. 2019, 70, 1299–1311. [Google Scholar] [CrossRef] [Green Version]
- Hu, W.; Han, Y.; Sheng, Y.; Wang, Y.; Pan, Q.; Nie, H. Mass spectrometry imaging for direct visualization of components in plant tissues. J. Sep. Sci. 2021, 44, 3462–3476. [Google Scholar] [CrossRef]
- Dong, Y.; Sonawane, P.; Cohen, H.; Polturak, G.; Feldberg, L.; Avivi, S.H.; Rogachev, I.; Aharoni, A. High mass resolution, spatial metabolite mapping enhances the current plant gene and pathway discovery toolbox. New Phytol. 2021, 228, 1986–2002. [Google Scholar] [CrossRef]
- Shanta, P.V.; Li, B.; Stuart, D.D.; Cheng, Q. Lipidomic profiling of algae with microarray MALDI-MS toward ecotoxicological monitoring of herbicide exposure. Environ. Sci. Technol. 2021, 55, 10558–10568. [Google Scholar] [CrossRef]
- Righetti, L.; Bhandari, D.R.; Rolli, E.; Tortorella, S.; Bruni, R.; Dall’Asta, C.; Spengler, B. Unveiling the spatial distribution of aflatoxin B1 and plant defense metabolites in maize using AP-SMALDI mass spectrometry imaging. Plant J. 2021, 106, 185–199. [Google Scholar] [CrossRef] [PubMed]
- Boughton, B.A.; Thinagaran, D. Mass spectrometry imaging (MSI) for plant metabolomics. Methods Mol. Biol. 2018, 1778, 241–252. [Google Scholar] [PubMed]
- Kulkarni, P.; Wilschut, R.A.; Verhoeven, K.J.F.; van der Putten, W.H.; Garbeva, P. LAESI mass spectrometry imaging as a tool to differentiate the root metabolome of native and range-expanding plant species. Planta 2018, 248, 1515–1523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Samarah, L.Z.; Khattar, R.; Tran, T.H.; Stopka, S.A.; Brantner, C.A.; Parlanti, P.; Veličković, D.; Shaw, J.B.; Agtuca, B.J.; Stacey, G.; et al. Single-cell metabolic profiling: Metabolite formulas from isotopic fine structures in heterogeneous plant cell populations. Anal. Chem. 2020, 92, 7289–7298. [Google Scholar] [CrossRef] [PubMed]
- Heavisides, E.; Rouger, C.; Reichel, A.; Ulrich, C.; Wenzel-Storjohann, A.; Sebens, S.; Tasdemir, D. Seasonal variations in the metabolome and bioactivity profile of Fucus vesiculosus extracted by an optimized, pressurized liquid extraction protocol. Mar. Drugs 2018, 16, 503. [Google Scholar] [CrossRef] [Green Version]
- Robinson, A.R.; Dauwe, R.; Mansfield, S.D. Assessing the between-background stability of metabolic effects arising from lignin-related transgenic modifications, in two Populus hybrids using non-targeted metabolomics. Tree Physiol. 2018, 38, 378–396. [Google Scholar] [CrossRef]
- Killiny, N.; Valim, M.F.; Jones, S.E.; Hijaz, F. Effect of different rootstocks on the leaf metabolite profile of ‘Sugar Belle’ mandarin hybrid. Plant Signal. Behav. 2018, 13, e1445934. [Google Scholar] [CrossRef]
- Lavergne, F.D.; Broeckling, C.D.; Cockrell, D.M.; Haley, S.D.; Peairs, F.B.; Jahn, C.E.; Heuberger, A.L. GC-MS Metabolomics to Evaluate the Composition of Plant Cuticular Waxes for Four Triticum aestivum Cultivars. Int. J. Mol. Sci. 2018, 9, 249. [Google Scholar] [CrossRef] [Green Version]
- Calumpang, C.L.F.; Saigo, T.; Watanabe, M.; Tohge, T. Cross-species comparison of fruit-metabolomics to elucidate metabolic regulation of fruit polyphenolics among Solanaceous crops. Metabolites 2020, 10, 209. [Google Scholar] [CrossRef]
- Fang, C.; Fernie, A.R.; Luo, J. Exploring the diversity of plant metabolism. Trends Plant Sci. 2019, 24, 83–98. [Google Scholar] [CrossRef]
- Razzaq, A.; Sadia, B.; Raza, A.; Khalid Hameed, M.; Saleem, F. Metabolomics: A Way Forward for Crop Improvement. Metabolites 2019, 9, 303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kroymann, J. Natural diversity and adaptation in plant secondary metabolism. Curr. Opin. Plant Biol. 2011, 14, 246–251. [Google Scholar] [CrossRef] [PubMed]
- Carrington, Y.; Guo, J.; Le, C.H.; Fillo, A.; Kwon, J.; Tran, L.T.; Ehlting, J. Evolution of a secondary metabolic pathway from primary metabolism: Shikimate and quinate biosynthesis in plants. Plant J. 2018, 95, 823–833. [Google Scholar] [CrossRef] [PubMed]
- Brunetti, C.; George, R.M.; Tattini, M.; Field, K.; Davey, M.P. Metabolomics in plant environmental physiology. J. Exp. Bot. 2013, 64, 4011–4020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Viant, M.R.; Sommer, U. Mass spectrometry based environmental metabolomics: A primer and review. Metabolomics 2013, 9, 144–158. [Google Scholar] [CrossRef]
- Littlejohn, G.R.; Breen, S.; Smirnoff, N.; Grant, M. Chloroplast immunity illuminated. New Phytol. 2021, 229, 3088–3107. [Google Scholar] [CrossRef]
- Garcia-Cela, E.; Kiaitsi, E.; Medina, A.; Sulyok, M.; Krska, R.; Magan, N. Interacting environmental stress factors affects targeted metabolomic profiles in stored natural wheat and that inoculated with F. graminearum. Toxins 2018, 10, 56. [Google Scholar] [CrossRef] [Green Version]
- Mills, G.; Sharps, K.; Simpson, D.; Pleijel, H.; Frei, M.; Burkey, K.; Emberson, L.; Uddling, J.; Broberg, M.; Feng, Z.; et al. Closing the global ozone yield gap: Quantification and CO benefits for multi stress tolerance. Glob. Change Biol. 2018, 24, 4869–4893. [Google Scholar] [CrossRef] [Green Version]
- Casatejada-Anchel, R.; Muñoz-Bertomeu, J.; Rosa-Téllez, S.; Anoman, A.D.; Nebauer, S.G.; Torres-Moncho, A.; Fernie, A.R.; Ros, R. Phosphoglycerate dehydrogenase genes differentially affect Arabidopsis metabolism and development. Plant Sci. 2021, 306, 110863. [Google Scholar] [CrossRef]
- Simpson, J.P.; Wunderlich, C.; Li, X.; Svedin, E.; Dilkes, B.; Chapple, C. Metabolic source isotopic pair labeling and genome-wide association are complementary tools for the identification of metabolite-gene associations in plants. Plant Cell 2021, 33, 492–510. [Google Scholar] [CrossRef]
- Salem, M.A.; Yoshida, T.; Perez de Souza, L.; Alseekh, S.; Bajdzienko, K.; Fernie, A.R.; Giavalisco, P. An improve extraction method enables the comprehensive analysis of lipids, proteins, metabolites and phytohormones from a single sample of leaf tissue under water-deficit stress. Plant J. 2020, 103, 1614–1632. [Google Scholar] [CrossRef] [PubMed]
- Serrano, N.; Ling, Y.; Bahieldin, A.; Mahfouz, M.M. Thermopriming reprograms metabolic homeostasis to confer heat tolerance. Sci. Rep. 2019, 9, 181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Küstner, L.; Fürtauer, L.; Weckwerth, W.; Nägele, T.; Heyer, A.G. Subcellular dynamics of proteins and metabolites under abiotic stress reveal deferred response of the Arabidopsis thaliana hexokinase-1 mutant gin2-1 to high light. Plant J. 2019, 100, 456–472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fürtauer, L.; Pschenitschnigg, A.; Scharkosi, H.; Weckwerth, W.; Nägele, T. Combined multivariate analysis and machine learning reveals a predictive module of metabolic stress response in Arabidopsis thaliana. Mol. Omics 2018, 14, 437–449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weiszmann, J.; Fürtauer, L.; Weckwerth, W.; Nägele, T. Vacuolar sucrose cleavage prevents limitation of cytosolic carbohydrate metabolism and stabilizes photosynthesis under abiotic stress. FEBS J. 2018, 285, 4082–4098. [Google Scholar] [CrossRef] [Green Version]
- David, L.; Kang, J.; Chen, S. targeted metabolomics of plant hormones and redox metabolites in stomatal immunity. Methods Mol. Biol. 2020, 2085, 79–92. [Google Scholar]
- Jurkiewicz, P.; Melser, S.; Maucourt, M.; Ayeb, H.; Veljanovski, V.; Maneta-Peyret, L.; Hooks, M.; Rolin, D.; Moreau, P.; Batoko, H. The multistress-induced Translocator protein (TSPO) differentially modulates storage lipids metabolism in seeds and seedlings. Plant J. 2018, 96, 274–286. [Google Scholar] [CrossRef]
- Georgii, E.; Jin, M.; Zhao, J.; Kanawati, B.; Schmitt-Kopplin, P.; Albert, A.; Winkler, J.B.; Schäffner, A.R. Relationships between drought, heat and air humidity responses revealed by transcriptome-metabolome co-analysis. BMC Plant Biol. 2017, 17, 120. [Google Scholar] [CrossRef] [Green Version]
- Ahkami, A.H.; Wang, W.; Wietsma, T.W.; Winkler, T.; Lange, I.; Jansson, C.; Lange, B.M.; McDowell, N.G. Metabolic shifts associated with drought-induced senescence in Brachypodium. Plant Sci. 2019, 289, 110278. [Google Scholar] [CrossRef]
- Hiraga, Y.; Shimada, N.; Nagashima, Y.; Suda, K.; Kanamori, T.; Ishiguro, K.; Sato, Y.; Hirakawa, H.; Sato, S.; Akashi, T.; et al. Identification of a Flavin Monooxygenase-Like Flavonoid 8-Hydroxylase with Gossypetin Synthase Activity from Lotus japonicus. Plant Cell Physiol. 2021, 62, 411–423. [Google Scholar] [CrossRef]
- Arif, M.A.; Alseekh, S.; Harb, J.; Fernie, A.; Frank, W. Abscisic acid, cold and salt stimulate conserved metabolic regulation in the moss Physcomitrella patens. Plant Biol. 2018, 20, 1014–1022. [Google Scholar] [CrossRef] [PubMed]
- Glaubitz, U.; Li, X.; Schaedel, S.; Erban, A.; Sulpice, R.; Kopka, J.; Hincha, D.K.; Zuther, E. Integrated analysis of rice transcriptomic and metabolomic responses to elevated night temperatures identifies sensitivity-and tolerance-related profiles. Plant Cell Environ. 2017, 40, 121–137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Auler, P.A.; Souza, G.M.; da Silva Engela, M.R.G.; do Amaral, M.N.; Rossatto, T.; da Silva, M.G.Z.; Furlan, C.M.; Maserti, B.; Braga, E.J.B. Stress memory of physiological, biochemical and metabolomic responses in two different rice genotypes under drought stress: The scale matters. Plant Sci. 2021, 311, 110994. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.; Cheong, B.E.; Natera, S.; Roessner, U. Morphological and metabolic responses to salt stress of rice (Oryza sativa L.) cultivars which differ in salinity tolerance. Plant Physiol. Biochem. 2019, 144, 427–435. [Google Scholar] [CrossRef] [PubMed]
- Sarabia, L.D.; Boughton, B.A.; Rupasinghe, T.; van de Meene, A.; Callahan, D.L.; Hill, C.B.; Roessner, U. High-mass-resolution MALDI mass spectrometry imaging reveals detailed spatial distribution of metabolites and lipids in roots of barley seedlings in response to salinity stress. Metab. Off. J. Metab. Soc. 2018, 14, 63. [Google Scholar] [CrossRef] [Green Version]
- Cao, D.; Lutz, A.; Hill, C.B.; Callahan, D.L.; Roessner, U. A quantitative profiling method of phytohormones and other metabolites applied to barley roots subjected to salinity stress. Front. Plant Sci. 2017, 7, 2070. [Google Scholar] [CrossRef] [Green Version]
- Xu, C.; Wei, L.; Huang, S.; Yang, C.; Wang, Y.; Yuan, H.; Xu, Q.; Zhang, W.; Wang, M.; Zeng, X.; et al. Drought resistance in Qingke involves a reprogramming of the phenylpropanoid pathway and UDP-glucosyltransferase regulation of abiotic stress tolerance targeting flavonoid biosynthesis. J. Agric. Food Chem. 2021, 69, 3992–4005. [Google Scholar] [CrossRef]
- Echeverria, A.; Larrainzar, E.; Li, W.; Watanabe, Y.; Sato, M.; Tran, C.D.; Moler, J.A.; Hirai, M.Y.; Sawada, Y.; Tran, L.P.; et al. Medicago sativa and Medicago truncatula show contrasting root metabolic responses to drought. Front. Plant Sci. 2021, 12, 652143. [Google Scholar] [CrossRef]
- Dickinson, E.; Rusilowicz, M.J.; Dickinson, M.; Charlton, A.J.; Bechtold, U.; Mullineaux, P.M.; Wilson, J. Integrating transcriptomic techniques and k-means clustering in metabolomics to identify markers of abiotic and biotic stress in Medicago truncatula. Metabolomics 2018, 14, 126. [Google Scholar] [CrossRef] [Green Version]
- Pagano, A.; de Sousa Araújo, S.; Macovei, A.; Dondi, D.; Lazzaroni, S.; Balestrazzi, A. Metabolic and gene expression hallmarks of seed germination uncovered by sodium butyrate in Medicago truncatula. Plant Cell Environ. 2019, 42, 259–269. [Google Scholar] [CrossRef] [Green Version]
- Pinasseau, L.; Vallverdú-Queralt, A.; Verbaere, A.; Roques, M.; Meudec, E.; Le Cunff, L.; Péros, J.P.; Ageorges, A. Cultivar Diversity of Grape Skin Polyphenol Composition and Changes in Response to Drought Investigated by LC-MS Based Metabolomics. Front. Plant Sci. 2017, 8, 1826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Furlan, A.L.; Bianucci, E.; Castro, S.; Dietz, K.J. Metabolic features involved in drought stress tolerance mechanisms in peanut nodules and their contribution to biological nitrogen fixation. Plant Sci. 2017, 263, 12–22. [Google Scholar] [CrossRef] [PubMed]
- Barding, G.A., Jr.; Béni, S.; Fukao, T.; Bailey-Serres, J.; Larive, C.K. Comparison of GC-MS and NMR for metabolite profiling of rice subjected to submergence stress. J. Proteome Res. 2012, 12, 98–909. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, S.; Wang, X.; Yin, X.; Nanjo, Y.; Ohyanagi, H.; Sakata, K. Integration of gel-based and gel-free proteomic data for functional analysis of proteins through soybean proteome database. J. Proteom. 2017, 163, 52–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herzog, M.; Fukao, T.; Winkel, A.; Konnerup, D.; Lamichhane, S.; Alpuerto, J.B.; Hasler-Sheetal, H.; Pedersen, O. Physiology, gene expression, and metabolome of two wheat cultivars with contrasting submergence tolerance. Plant Cell Environ. 2018, 41, 1632–1644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luengwilai, K.; Saltveit, M.; Beckles, D.M. Metabolite content of harvested micro-tom tomato (Solanum lycopersicum L.) fruit is altered by chilling and protective heat-shock treatments as shown by GC–MS metabolic profiling. Postharvest. Biol. Technol. 2012, 63, 116–122. [Google Scholar] [CrossRef]
- Thomason, K.; Babar, M.A.; Erickson, J.E.; Mulvaney, M.; Beecher, C.; MacDonald, G. Comparative physiological and metabolomics analysis of wheat (Triticum aestivum L.) following post-anthesis heat stress. PLoS ONE 2018, 13, e0197919. [Google Scholar]
- Silvente, S.; Sobolev, A.P.; Lara, M. Metabolite adjustments in drought tolerant and sensitive soybean genotypes in response to water stress. PLoS ONE 2012, 7, e38554. [Google Scholar] [CrossRef] [Green Version]
- Chebrolu, K.K.; Fritschi, F.B.; Ye, S.; Krishnan, H.B.; Smith, J.R.; Gillman, J.D. Impact of heat stress during seed development on soybean seed metabolome. Metabolomics 2016, 12, 28. [Google Scholar] [CrossRef]
- de Oliveira, D.F.; Lopes, L.D.; Gomes-Filho, E. Metabolic changes associated with differential salt tolerance in sorghum genotypes. Planta 2020, 252, 34. [Google Scholar] [CrossRef]
- Paidi, M.K.; Agarwal, P.; More, P.; Agarwal, P.K. Chemical derivatization of metabolite mass profiling of the recretohalophyte Aeluropus lagopoides revealing salt stress tolerance mechanism. Mar. Biotechnol. 2017, 19, 207–218. [Google Scholar] [CrossRef] [PubMed]
- Younessi-Hamzekhanlu, M.; Dibazarnia, Z.; Oustan, S.; Vinson, T.; Katam, R.; Mahna, N. Mild salinity stimulates biochemical activities and metabolites associated with anticancer activities in black horehound (Ballota nigra L.). Agronomy 2021, 11, 2538. [Google Scholar] [CrossRef]
- Benjamin, J.J.; Lucini, L.; Jothiramshekar, S.; Parida, A. Metabolomic insights into the mechanisms underlying tolerance to salinity in different halophytes. Plant Physiol. Biochem. 2019, 135, 528–545. [Google Scholar] [CrossRef] [PubMed]
- Jahangir, M.; Abdel-Farid, I.B.; Choi, Y.H.; Verpoorte, R. Metal ion-inducing metabolite accumulation in Brassica rapa. J. Plant Physiol. 2008, 165, 1429–1437. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez Ibarra, A.A.; Wrobel, K.; Barrientos, E.Y.; Corrales Escobosa, A.R.; Gutierrez Corona, J.F.; Donis, I.E.; Wrobel, K. Changes of metabolomic profile in Helianthus annuus under exposure to chromium (VI) studied by HPLC-ESI-QTOF-MS and MS/MS. J. Anal. Methods Chem. 2017, 2017, 3568621. [Google Scholar] [CrossRef] [Green Version]
- Pott, D.M.; de Abreu, E.; Lima, F.; Soria, C.; Willmitzer, L.; Fernie, A.R.; Nikoloski, Z.; Osorio, S.; Vallarino, J.G. Metabolic reconfiguration of strawberry physiology in response to postharvest practices. Food Chem. 2020, 321, 126747. [Google Scholar] [CrossRef]
- Sung, J.; Lee, S.; Lee, Y.; Ha, S.; Song, B.; Kim, T.; Waters, B.M.; Krishnan, H.B. Metabolomic profiling from leaves and roots of tomato (Solanum lycopersicum L.) plants grown under nitrogen, phosphorus or potassium-deficient condition. Plant Sci. 2015, 241, 55–64. [Google Scholar] [CrossRef]
- Ghosson, H.; Schwarzenberg, A.; Jamois, F.; Yvin, J.C. Simultaneous untargeted and targeted metabolomics profiling of underivatized primary metabolites in sulfur-deficient barley by ultra-high performance liquid chromatography-quadrupole/time-of-flight mass spectrometry. Plant Methods 2018, 14, 62. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Ma, X.M.; Wang, X.C.; Liu, J.H.; Huang, B.Y.; Guo, X.Y.; Xiong, S.P.; La, G.X. UPLC-QTOF analysis reveals metabolomic changes in the flag leaf of wheat (Triticum aestivum L.) under low-nitrogen stress. Plant Physiol. Biochem. 2017, 111, 30–38. [Google Scholar] [CrossRef]
- Sheflin, A.M.; Chiniquy, D.; Yuan, C.; Goren, E.; Kumar, I.; Braud, M.; Brutnell, T.; Eveland, A.L.; Tringe, S.; Liu, P.; et al. Metabolomics of sorghum roots during nitrogen stress reveals compromised metabolic capacity for salicylic acid biosynthesis. Plant Direct. 2019, 3, e00122. [Google Scholar] [CrossRef] [Green Version]
- Tugizimana, F.; Mhlongo, M.I.; Piater, L.A.; Dubery, I.A. Metabolomics in plant priming research: The way forward? Int. J. Mol. Sci. 2018, 19, 1759. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vinci, G.; Cozzolino, V.; Mazzei, P.; Monda, H.; Spaccini, R.; Piccolo, A. An alternative to mineral phosphorus fertilizers: The combined effects of Trichoderma harzianum and compost on Zea mays, as revealed by 1H NMR and GC-MS metabolomics. PLoS ONE 2018, 13, e0209664. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miotto-Vilanova, L.; Courteaux, B.; Padilla, R.; Rabenoelina, F.; Jacquard, C.; Clément, C.; Comte, G.; Lavire, C.; AitBarka, E.; Kerzaon, I.; et al. Impact of Paraburkholderia phytofirmans PsJN on grapevine phenolic metabolism. Int. J. Mol. Sci. 2019, 20, 5775. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brader, G.; Company, S.; Mitter, B.; Trognitz, F.; Sessitsch, A. Metabolic potential of endophytic bacteria. Curr. Opin. Biotechnol. 2014, 27, 30–37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sasse, J.; Martinoia, E.; Northen, T. Feed your friends: Do plant exudates shape the root microbiome? Trends Plant Sci. 2017, 23, 25–41. [Google Scholar] [CrossRef] [Green Version]
- Cotton, T.E.A.; Petriacq, P.; Cameron, D.D.; Meselmani, M.A.; Schwarzenbacher, R.; Rolfe, S.A.; Ton, J. Metabolic regulation of the maize rhizobiome by benzoxazinoids. ISME J. 2019, 13, 1647–1658. [Google Scholar] [CrossRef] [Green Version]
- Zhalnina, K.; Louie, K.B.; Hao, Z.; Mansoori, N.; da Rocha, U.N.; Shi, S.; Cho, H.; Karaoz, U.; Loqué, D.; Bowen, B.P.; et al. Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat. Microbiol. 2018, 3, 470–480. [Google Scholar] [CrossRef] [Green Version]
- Abd El-Daim, I.A.; Bejai, S.; Meijer, J. Bacillus velezensis 5113 induced metabolic and molecular reprogramming during abiotic stress tolerance in wheat. Sci. Rep. 2019, 9, 16282. [Google Scholar] [CrossRef] [Green Version]
- Cherif-Silini, H.; Thissera, B.; Bouket, A.C.; Saadaoui, N.; Silini, A.; Eshelli, M.; Alenezi, F.N.; Vallat, A.; Luptakova, L.; Yahiaoui, B.; et al. Durum wheat stress tolerance induced by endophyte Pantoea agglomerans with genes contributing to plant functions and secondary metabolite arsenal. Int. J. Mol. Sci. 2019, 20, 3989. [Google Scholar] [CrossRef] [Green Version]
- Pang, Q.; Zhang, T.; Wang, Y.; Kong, W.; Guan, Q.; Yan, X.; Chen, S. Metabolomics of early stage plant cell-microbe interaction using stable isotope labeling. Front. Plant Metab. Chemodiversity 2018, 9, 760. [Google Scholar] [CrossRef] [Green Version]
- Papantoniou, D.; Vergara, F.; Weinhold, A.; Quijano, T.; Khakimov, B.; Pattison, D.I.; Bak, S.; van Dam, N.M.; Martínez-Medina, A. Cascading effects of root microbial symbiosis on the development and metabolome of the insect herbivore Manduca sexta L. Metabolites 2021, 11, 731. [Google Scholar] [CrossRef] [PubMed]
- Richards, L.A.; Glassmire, A.E.; Ochsenrider, K.M.; Smilanich, A.M.; Dodson, C.D.; Jeffrey, C.S.; Dyer, L.A. Phytochemical diversity and synergistic effects on herbivores. Phytochem. Rev. 2016, 15, 1153–1166. [Google Scholar] [CrossRef]
- Kovalikova, Z.; Kubes, J.; Skalicky, M.; Kuchtickova, N.; Maskova, L.; Tuma, J.; Vachova, P.; Hejnak, V. Changes in content of polyphenols and ascorbic acid in leaves of white cabbage after pest infestation. Molecules 2019, 24, 2622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kariyat, R.R.; Gaffoor, I.; Sattar, S.; Dixon, C.W.; Frock, N.; Moen, J.; De Moraes, C.M.; Mescher, M.C.; Thompson, G.A.; Chopra, S. Sorghum 3-Deoxyanthocyanidin flavonoids confer resistance against corn leaf aphid. J. Chem. Ecol. 2019, 45, 502–514. [Google Scholar] [CrossRef]
- Tayal, M.; Somavat, P.; Rodriguez, I.; Martinez, L.; Kariyat, R. Cascading effects of polyphenol-rich purple corn pericarp extract on pupal, adult, and offspring of tobacco hornworm (Manduca sexta L.). Commun. Integr. Biol. 2020, 13, 43–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chhajed, S.; Mostafa, I.; He, Y.; Abou-Hashem, M.; El-Domiaty, M.; Chen, S. Glucosinolate biosynthesis and the glucosinolate–myrosinase system in plant defense. Agronomy 2020, 10, 1786. [Google Scholar] [CrossRef]
- Machado, R.A.R.; Theepan, V.; Robert, C.A.M.; Züst, T.; Hu, L.; Su, Q.; Schimmel, B.C.J.; Erb, M. The plant metabolome guides fitness-relevant foraging decisions of a specialist herbivore. PLoS Biol. 2021, 19, e3001114. [Google Scholar] [CrossRef]
- Macel, M.; Visschers, I.G.S.; Peters, J.L.; Kappers, I.F.; de Vos, R.C.H.; van Dam, N.M. Metabolomics of thrips resistance in pepper (Capsicum spp.) reveals monomer and dimer acyclic diterpene glycosides as potential chemical defenses. J. Chem. Ecol. 2019, 45, 490–501. [Google Scholar] [CrossRef] [Green Version]
- Aerts, N.; Pereira Mendes, M.; Van Wees, S.C.M. Multiple levels of crosstalk in hormone networks regulating plant defense. Plant J. 2021, 105, 489–504. [Google Scholar] [CrossRef]
- Munné-Bosch, S.; Müller, M. Hormonal cross-talk in plant development and stress responses. Front. Plant Sci. 2013, 4, 529. [Google Scholar] [CrossRef] [Green Version]
- Lin, C.; Lott, A.A.; Zhu, W.; Dufresne, C.P.; Chen, S. Mitogen-activated protein kinase 4-regulated metabolic networks. Int. J. Mol. Sci. 2022, 23, 880. [Google Scholar] [CrossRef] [PubMed]
- Hildreth, S.B.; Foley, E.E.; Muday, G.K.; Helm, R.F.; Winkel, B.S.J. The dynamic response of the Arabidopsis root metabolome to auxin and ethylene is not predicted by changes in the transcriptome. Sci. Rep. 2020, 10, 679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, X.; Wang, R.S.; Zhu, M.; Jeon, B.W.; Albert, R.; Chen, S.; Assmann, S.M. Abscisic acid-responsive guard cell metabolomes of Arabidopsis wild-type and gpa1 G-protein mutants. Plant Cell. 2013, 25, 4789–4811. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yousaf, M.J.; Hussain, A.; Hamayun, M.; Iqbal, A.; Irshad, M.; Kim, H.-Y.; Lee, I.-J. Transformation of Endophytic bipolaris spp. into biotrophic pathogen under auxin cross-talk with brassinosteroids and abscisic acid. Front Bioeng. Biotechnol. 2021, 9, 657635. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Min, C.W.; Kramer, K.; Agrawal, G.K.; Rakwal, R.; Park, K.-H.; Wang, Y.; Finkemeier, I.; Kim, S.T. A multi-omics analysis of Glycine max leaves reveals alteration in flavonoid and isoflavonoid metabolism upon ethylene and abscisic acid treatment. Proteomics 2018, 18, e1700366. [Google Scholar] [CrossRef]
- Wang, J.F.; Liu, S.S.; Song, Z.Q.; Xu, T.C.; Liu, C.S.; Hou, Y.G.; Huang, R.; Wu, S.H. Naturally Occurring Flavonoids and Isoflavonoids and Their Microbial Transformation: A Review. Molecules 2020, 25, 5112. [Google Scholar] [CrossRef]
- Lerner, A.B.; Case, J.D.; Takahashi, Y.; Lee, T.H.; Mori, W. Isolation of melatonin, a pineal factor that lightens melanocytes. J. Am. Chem. Soc. 1958, 80, 2587. [Google Scholar] [CrossRef]
- Arnao, M.B.; Hernandez-Ruiz, J. Melatonin as a regulatory hub of plant hormone levels and action in stress situations. Plant Biol. 2021, 23, 7–19. [Google Scholar] [CrossRef]
- Sun, C.; Liu, L.; Wang, L.; Li, B.; Jin, C.; Lin, X. Melatonin: A master regulator of plant development and stress responses. J. Integr. Plant Biol. 2020, 63, 126–145. [Google Scholar] [CrossRef]
- Xie, Z.; Wang, J.; Wang, W.; Wang, Y.; Xu, J.; Li, Z.; Zhao, X.; Fu, B. Integrated Analysis of the Transcriptome and Metabolome Revealed the Molecular Mechanisms Underlying the Enhanced Salt Tolerance of Rice Due to the Application of Exogenous Melatonin. Front. Plant Sci. 2021, 11, 618680. [Google Scholar] [CrossRef]
- Arnao, M.B.; Hernández-Ruiz, J. Melatonin: A new plant hormone and/or a plant master regulator? Trends Plant Sci. 2019, 24, 38–48. [Google Scholar] [CrossRef] [PubMed]
- Tan, D.-X.; Reiter, R.J. An evolutionary view of melatonin synthesis and metabolism related to its biological functions in plants. J. Exp. Bot. 2020, 71, 4677–4689. [Google Scholar] [CrossRef] [PubMed]
- Bernal-Vicente, A.; Cantabella, D.; Petri, C.; Hernández, J.A.; Diaz-Vivancos, P. The salt-stress response of the transgenic plum line J8-1 and its interaction with the salicylic acid biosynthetic pathway from mandelonitrile. Int. J. Mol. Sci. 2018, 19, 3519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lima, M.R.M.; Machado, A.F.; Gubler, W.D. Metabolomic Study of Chardonnay Grapevines Double Stressed with Esca-Associated Fungi and Drought. Phytopathology 2017, 107, 669–680. [Google Scholar] [CrossRef] [PubMed]
- Ueno, V.A.; Sawaya, A. Influence of environmental factors on the volatile composition of two Brazilian medicinal plants: Mikania laevigata and Mikania glomerata. Metabolomics 2019, 15, 91. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Kwon, M.C.; Jung, E.S.; Lee, C.H.; Oh, M.M. Physiological and metabolomic responses of kale to combined chilling and UV-A treatment. Int. J. Mol. Sci. 2019, 20, 4950. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Xing, H.; Li, X.; Geng, S.; Ning, D.; Ma, T.; Yu, X. Physiological and metabolomics analyses reveal the roles of fulvic acid in enhancing the production of astaxanthin and lipids in Haematococcus pluvialis under abiotic stress conditions. J. Agric. Food Chem. 2019, 67, 12599–12609. [Google Scholar] [CrossRef]
- Casartelli, A.; Melino, V.J.; Baumann, U.; Riboni, M.; Suchecki, R.; Jayasinghe, N.S.; Mendis, H.; Watanabe, M.; Erban, A.; Zuther, E.; et al. Opposite fates of the purine metabolite allantoin under water and nitrogen limitations in bread wheat. Plant Mol. Biol 2019, 99, 477–497. [Google Scholar] [CrossRef] [Green Version]
- Caldana, C.; Degenkolbe, T.; Cuadros-Inostroza, A.; Klie, S.; Sulpice, R.; Leisse, A.; Steinhauser, D.; Fernie, A.R.; Willmitzer, L.; Hannah, M. High-density kinetic analysis of the metabolomic and transcriptomic response of Arabidopsis to eight environmental conditions. Plant J. 2011, 67, 869–884. [Google Scholar] [CrossRef]
- Fraire-Velázquez, S.; Balderas-Hernández, V.E. Abiotic Stress in Plants and Metabolic Responses. In Abiotic Stress—Plant Responses and Applications in Agriculture, 1st ed.; Vahdati, K., Leslie, C., Eds.; Intechopen: London, UK, 2013; Volume 1, 420p. [Google Scholar]
- Schwachtje, J.; Whitcomb, S.J.; Firmino, A.A.P.; Zuther, E.; Hincha, D.K.; Kopka, J. Induced, imprinted, and primed responses to changing environments: Does metabolism store and process information? Front. Plant Sci. 2019, 10, 106. [Google Scholar] [CrossRef]
- Murcia, G.; Fontana, A.; Pontin, M.; Baraldi, R.; Bertazza, G.; Piccoli, P.N. ABA and GA3 regulate the synthesis of primary and secondary metabolites related to alleviation from biotic and abiotic stresses in grapevine. Phytochemistry 2017, 135, 34–52. [Google Scholar] [CrossRef] [PubMed]
- Nishanth, M.J.; Sheshadri, S.A.; Rathore, S.S.; Srinidhi, S.; Simon, B. Expression analysis of cell wall invertase under abiotic stress conditions influencing specialized metabolism in Catharanthus roseu. Sci. Rep. 2018, 8, 15059. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lucini, L.; Rouphael, Y.; Cardarelli, M.; Bonini, P.; Baffi, C.; Colla, G. A vegetal biopolymer-based biostimulant promoted root growth in melon while triggering brassinosteroids and stress-related compounds. Front. Plant Sci. 2018, 9, 472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bird, A. Perceptions of epigenetics. Nature 2007, 447, 396–398. [Google Scholar] [CrossRef]
- Mach, J. Identification of a novel maize protein important for paramutation at the purple plant1 locus. Plant Cell 2012, 24, 1709. [Google Scholar] [CrossRef] [Green Version]
- Kumar, R.; Abhishek, B.; Pandey, A.K.; Pandey, M.K.; Kumar, A. Metabolomics for plant improvement: Status and prospects. Front. Plant Sci. 2017, 8, 1302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manning, K.; Tör, M.; Poole, M.; Hong, Y.; Thompson, A.J.; King, G.J.; Giovannoni, J.J.; Seymour, G.B. A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat Genet. 2006, 38, 948–952. [Google Scholar] [CrossRef]
- Osorio, S.; Alba, R.; Damasceno, C.M.; Lopez-Casado, G.; Lohse, M.; Zanor, M.I.; Fernie, A.R. Systems biology of tomato fruit development: Combined transcript, protein, and metabolite analysis of tomato transcription factor (nor, rin) and ethylene receptor (Nr) mutants reveals novel regulatory interactions. Plant Physiol. 2011, 157, 405–425. [Google Scholar] [CrossRef] [Green Version]
- Lazcano-Ramírez, H.; Gamboa-Becerra, R.; García-López, I.; Montes, R.; Díaz-Ramírez, D.; Vega, O.; Ordaz-Ortíz, J.; de Folter, S.; Tiessen-Favier, A.; Winkler, R.; et al. Effects of the developmental regulator bolita on the plant metabolome. Genes 2021, 12, 995. [Google Scholar] [CrossRef]
- Fondi, M.; Liò, P. Multi-omics and metabolic modelling pipelines: Challenges and tools for systems microbiology. Microbiol. Res. 2015, 171, 52–64. [Google Scholar] [CrossRef]
- Wen, S.; Li, C.; Zhan, X. Muti-omics integration analysis revealed molecular network alterations in human nonfunctional pituitary neuroendocrine tumors in the framework of 3P medicine. EPMA J. 2022, 13, 9–37. [Google Scholar] [CrossRef] [PubMed]
- Shetty, S.A.; Smidt, H.; de Vos, W.M. Reconstructing functional networks in the human intestinal tract using synthetic microbiomes. Curr. Opin. Biotechnol. 2019, 58, 146–154. [Google Scholar] [CrossRef] [PubMed]
- García-Sevillano, M.Á.; Garcia-Barrera, T.; Abril, N.; Pueyo, C.; Lopez-Barea, J.; Gómez-Ariza, J.L. Omics technologies and their applications to evaluate metal toxicity in mice M. spretus as a bioindicator. J. Proteom. 2014, 104, 4–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gutleben, J.; Chai De Mares, M.; van Elsas, J.D.; Smidt, H.; Overmann, J.; Sipkema, D. The multi-omics promise in context: From sequence to microbial isolate. Crit. Rev. Microbiol. 2018, 44, 212–229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X. Protein and proteome atlas for plants under stresses: New highlights and ways for integrated omics in post-genomics era. Int. J. Mol. Sci. 2019, 20, 5222. [Google Scholar] [CrossRef] [Green Version]
- Gai, Z.; Wang, Y.; Ding, Y.; Qian, W.; Qiu, C.; Xie, H.; Sun, L.; Jiang, Z.; Ma, Q.; Wang, L.; et al. Exogenous abscisic acid induces the lipid and flavonoid metabolism of tea plants under drought stress. Sci. Rep. 2020, 10, 12275. [Google Scholar] [CrossRef]
- Yu, A.; Zhao, J.; Wang, Z.; Cheng, K.; Zhang, P.; Tian, G.; Liu, X.; Guo, E.; Du, Y.; Wang, Y. Transcriptome and metabolite analysis reveal the drought tolerance of foxtail millet significantly correlated with phenylpropanoids-related pathways during germination process under PEG stress. BMC Plant Biol. 2020, 20, 274. [Google Scholar] [CrossRef]
- You, J.; Zhang, Y.; Liu, A.; Li, D.; Wang, X.; Dossa, K.; Zhou, R.; Yu, J.; Zhang, Y.; Wang, L.; et al. Transcriptomic and metabolomic profiling of drought-tolerant and susceptible sesame genotypes in response to drought stress. BMC Plant Biol. 2019, 19, 267. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Ma, K.-B.; Lu, Z.-G.; Ren, S.-X.; Jiang, H.-R.; Cui, J.-W.; Chen, G.; Teng, N.-J.; Lam, H.-M.; Jin, B. Differential physiological, transcriptomic and metabolomic responses of Arabidopsis leaves under prolonged warming and heat shock. BMC Plant Biol. 2020, 20, 86. [Google Scholar] [CrossRef]
- Alhaithloul, H.A.; Soliman, M.H.; Ameta, K.L.; El-Esawi, M.A.; Elkelish, A. Changes in ecophysiology, osmolytes, and secondary metabolites of the medicinal plants of Mentha piperita and Catharanthus roseus subjected to drought and heat stress. Biomolecules 2020, 10, 43. [Google Scholar] [CrossRef] [Green Version]
- Natarajan, P.; Akinmoju, T.A.; Nimmakayala, P.; Lopez-Ortiz, C.; Garcia-Lozano, M.; Thompson, B.J.; Stommel, J.; Reddy, U.K. Integrated metabolomic and transcriptomic analysis to characterize cutin biosynthesis between low-and high-cutin genotypes of Capsicum chinense Jacq. Int. J. Mol. Sci. 2020, 21, 1397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Jiang, X.; Zhao, C.; Fang, Z.; Jiao, P. Transcriptomic and metabolomic analysis reveals the role of CoA in the salt tolerance of Zygophyllum spp. BMC Plant Biol. 2020, 20, 9. [Google Scholar] [CrossRef] [PubMed]
- Pi, E.; Xu, J.; Li, H.; Fan, W.; Zhu, C.; Zhang, T.; Jiang, J.; He, L.; Lu, H.; Wang, H.; et al. Enhanced salt tolerance of rhizobia-inoculated soybean correlates with decreased phosphorylation of the transcription factor GmMYB183 and altered flavonoid biosynthesis. Mol. Cell. Proteom. 2019, 18, 2225–2243. [Google Scholar] [CrossRef] [PubMed]
- Niu, Y.; Liu, Z.; He, H.; Han, X.; Qi, Z.; Yang, Y. Gene expression and metabolic changes of Momordica charantia L. seedlings in response to low temperature stress. PLoS ONE 2020, 15, e0233130. [Google Scholar] [CrossRef] [PubMed]
- Hildebrandt, T.M. Synthesis versus degradation: Directions of amino acid metabolism during Arabidopsis abiotic stress response. Plant Mol. Biol. 2018, 98, 121–135. [Google Scholar] [CrossRef] [PubMed]
- Katam, R.; Shokri, S.; Murthy, N.; Singh, S.K.; Suravajhala, P.; Khan, M.N.; Bahmani, M.; Sakata, K.; Reddy, K.R. Proteomics, physiological, and biochemical analysis of cross tolerance mechanisms in response to heat and water stresses in soybean. PLoS ONE 2020, 15, e0233905. [Google Scholar] [CrossRef]
- Ghareeb, H.; El-Sayed, M.; Pound, M.; Tetyuk, O.; Hanika, K.; Herrfurth, C.; Feussner, I.; Lipka, V. Quantitative hormone signaling output analyses of Arabidopsis thaliana interactions with virulent and avirulent Hyaloperonospora arabidopsidis isolates at single-cell resolution. Front. Plant Sci. 2020, 11, 603693. [Google Scholar] [CrossRef]
- Mubeen, U.; Giraldi, L.A.; Jüppner, J.; Giavalisco, P. A Multi-omics extraction method for the in-depth analysis of synchronized cultures of the green alga Chlamydomonas reinhardtii. J. Vis. Exp. 2019, 150, 59547. [Google Scholar] [CrossRef] [Green Version]
- Cahill, J.F.; Kertesz, V. Laser capture microdissection—Liquid vortex capture mass spectrometry metabolic profiling of single onion epidermis and microalgae cells. Methods Mol. Biol. 2020, 2064, 89–101. [Google Scholar]
- Wada, H.; Chang, F.-Y.; Hatakeyama, Y.; Erra-Balsells, R.; Araki, T.; Nakano, H.; Nonami, H. Endosperm cell size reduction caused by osmotic adjustment during nighttime warming in rice. Sci. Rep. 2021, 11, 4447. [Google Scholar] [CrossRef]
- Ullah, M.A.; Tungmunnithum, D.; Garros, L.; Drouet, S.; Hano, C.; Abbasi, B.H. Effect of ultraviolet-c radiation and melatonin stress on biosynthesis of antioxidant and antidiabetic metabolites produced in in vitro callus cultures of Lepidium sativum L. Int. J. Mol. Sci. 2019, 20, 1787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kelley, Z.D.; Rogers, D.T.; Littleton, J.M.; Lynn, B.C. Microfluidic capillary zone electrophoresis mass spectrometry analysis of alkaloids in Lobelia cardinalis transgenic and mutant plant cell cultures. Electrophoresis 2019, 40, 2921–2928. [Google Scholar] [CrossRef] [PubMed]
- Wahyuni, D.K.; Rahayu, S.; Zaidan, A.H.; Ekasari, W.; Prasongsuk, S.; Purnobasuki, H. Growth, secondary metabolite production, and in vitro antiplasmodial activity of Sonchus arvensis L. callus under dolomite [CaMg (CO3)2] treatment. PLoS ONE 2021, 16, e0254804. [Google Scholar] [CrossRef] [PubMed]
- Shao, Q.; Hu, L.; Qin, H.; Liu, Y.; Tang, X.; Lei, A.; Wang, J. Metabolomic response of Euglena gracilis and its bleached mutant strain to light. PLoS ONE 2019, 14, e0224926. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Wang, H.P.; Yu, C.; Ding, W.; Han, B.; Geng, S.; Ning, D.; Ma, T.; Yu, X. Integration of physiological and metabolomic profiles to elucidate the regulatory mechanisms underlying the stimulatory effect of melatonin on astaxanthin and lipids coproduction in Haematococcus pluvialis under inductive stress conditions. Bioresour. Technol. 2021, 319, 124150. [Google Scholar] [CrossRef] [PubMed]
- Baumeister, T.U.H.; Vallet, M.; Kaftan, F.; Svatoš, A.; Pohnert, G. Live Single-cell metabolomics with matrix-free laser/desorption ionization mass spectrometry to address microalgal physiology. Front. Plant Sci. 2019, 10, 172. [Google Scholar] [CrossRef]
- Rippin, M.; Pichrtová, M.; Arc, E.; Kranner, I.; Becker, B.; Holzinger, A. Metatranscriptomic and metabolite profiling reveals vertical heterogeneity within a Zygnema green algal mat from Svalbard (High Arctic). Environ. Microbiol. 2019, 21, 4283–4299. [Google Scholar] [CrossRef] [Green Version]
- Yamada, K.; Nakanowatari, M.; Yumoto, E.; Satoh, S.; Asahina, M. Spatiotemporal plant hormone analysis from cryosections using laser microdissection-liquid chromatography-mass spectrometry. J. Plant Res. 2022, 135, 377–386. [Google Scholar] [CrossRef]
- Hieta, J.P.; Sipari, N.; Räikkönen, H.; Keinänen, M.; Kostiainen, R. Mass spectrometry imaging of Arabidopsis thaliana leaves at the single-cell level by infrared laser ablation atmospheric pressure photoionization (LAAPPI). J. Am. Soc. Mass Spectrom. 2021, 32, 2895–2903. [Google Scholar] [CrossRef]
- Sun, L.; Wang, R.; Ju, Q.; Xu, J. Physiological, metabolic, and transcriptomic analyses reveal the responses of Arabidopsis seedlings to carbon nanohorns. Environ. Sci. Technol. 2020, 54, 4409–4420. [Google Scholar] [CrossRef]
- Launay, A.; Cabassa-Hourton, C.; Eubel, H.; Maldiney, R.; Guivarc’h, A.; Crilat, E.; Savouré, A. Proline oxidation fuels mitochondrial respiration during dark-induced leaf senescence in Arabidopsis thaliana. J. Exp. Bot. 2019, 70, 6203–6214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taylor, M.J.; Mattson, S.; Liyu, A.; Stopka, S.A.; Ibrahim, Y.M.; Vertes, A.; Anderton, C.R. Optical Microscopy-Guided Laser Ablation Electrospray Ionization Ion Mobility Mass Spectrometry: Ambient Single Cell Metabolomics with Increased Confidence in Molecular Identification. Metabolites 2021, 11, 200. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Fan, Y.; Zhang, L.; Zheng, C.; Chen, A.; Sun, Y.; Guo, H.; Wu, J.; Li, T.; Fan, Y.; et al. Quantitative metabolome and transcriptome analysis reveals complex regulatory pathway underlying photoinduced fiber color formation in cotton. Gene 2021, 767, 145180. [Google Scholar] [CrossRef] [PubMed]
- Blokhina, O.; Laitinen, T.; Hatakeyama, Y.; Delhomme, N.; Paasela, T.; Zhao, L.; Fagerstedt, K. Parenchymal cells contribute to lignification of tracheids in developing xylem of norway spruce. Plant Physiol. 2019, 181, 1552–1572. [Google Scholar] [CrossRef] [Green Version]
- Figueiredo, J.; Cavaco, A.R.; Guerra-Guimarães, L.; Leclercq, C.; Renaut, J.; Cunha, J.; Eiras-Dias, J.; Cordeiro, C.; Matos, A.R.; Silva, M.S.; et al. An apoplastic fluid extraction method for the characterization of grapevine leaves proteome and metabolome from a single sample. Physiol. Plant. 2021, 171, 343–357. [Google Scholar] [CrossRef] [PubMed]
- Dueñas, M.E.; Larson, E.A.; Lee, Y.J. Toward mass spectrometry imaging in the metabolomics scale: Increasing metabolic coverage through multiple on-tissue chemical modifications. Front. Plant Sci. 2019, 10, 860. [Google Scholar] [CrossRef] [Green Version]
- DeHoog, R.J.; Zhang, J.; Alore, E.; Lin, J.Q.; Yu, W.; Woody, S.; Almendariz, C.; Lin, M.; Engelsman, A.F.; Sidhu, S.B.; et al. Preoperative metabolic classification of thyroid nodules using mass spectrometry imaging of fine-needle aspiration biopsies. Proc. Natl. Acad. Sci. USA 2019, 116, 21401–21408. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Cheng, S.; Lin, Q.; Cao, W.; Yang, J.; Zhang, M.; Shen, A.; Zhang, W.; Xia, Y.; Ma, X.; et al. Single-cell lipidomics with high structural specificity by mass spectrometry. Nat. Commun. 2021, 12, 2869. [Google Scholar] [CrossRef]
- You, W.; Wei, L.; Gong, Y.; Hajjami, M.E.; Xu, J.; Poetsch, A. Integration of proteome and transcriptome refines key molecular processes underlying oil production in Nannochloropsis oceanica. Biotechnol. Biofuels 2020, 13, 109. [Google Scholar] [CrossRef]
- Dai, W.; Hu, Z.; Xie, D.; Tan, J.; Lin, Z. A novel spatial-resolution targeted metabolomics method in a single leaf of the tea plant (Camellia sinensis). Food Chem. 2020, 311, 126007. [Google Scholar] [CrossRef]
- Lee, J.; Hyeon, D.Y.; Hwang, D. Single-cell multiomics: Technologies and data analysis methods. Exp. Mol. Med. 2020, 52, 1428–1442. [Google Scholar] [CrossRef] [PubMed]
- Thibivilliers, S.; Libault, M. Enhancing our understanding of plant cell-to-cell interactions using single-cell omics. Front. Plant Sci. 2021, 12, 696811. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, P.; Rasool, S.; Gul, A.; Sheikh, S.A.; Akram, N.A.; Ashraf, M.; Gucel, S. Jasmonates: Multifunctional roles in stress tolerance. Front. Plant Sci. 2016, 7, 813. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Priya, M.; Dhanker, O.P.; Siddique, K.H.M.; HanumanthaRao, B.; Nair, R.M.; Pandey, S.; Singh, S.; Varshney, R.K.; Prasad, P.V.V.; Nayyar, H. Drought and heat stress-related proteins: An update about their functional relevance in imparting stress tolerance in agricultural crops. Theor. Appl. Genet. 2019, 132, 1607–1638. [Google Scholar] [CrossRef]
- Ma, S.; Lv, L.; Meng, C.; Zhang, C.; Li, Y. Integrative Analysis of the Metabolome and Transcriptome of Sorghum bicolor Reveals Dynamic Changes in Flavonoids Accumulation under Saline–Alkali Stress. J. Agric. Food Chem. 2020, 68, 14781–14789. [Google Scholar] [CrossRef] [PubMed]
- Kang, K.B.; Ernst, M.; van der Hooft, J.J.J.; da Silva, R.R.; Park, J.; Medema, M.H.; Sung, S.H.; Dorrestein, P.C. Comprehensive mass spectrometry-guided phenotyping of plant specialized metabolites reveals metabolic diversity in the cosmopolitan plant family Rhamnaceae. Plant J. 2019, 98, 1134–1144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, K.; Logacheva, M.D.; Meng, Y.; Hu, J.; Wan, D.; Li, L.; Janovská, D.; Wang, Z.; Georgiev, M.I.; Yu, Z.; et al. Jasmonate-responsive MYB factors spatially repress rutin biosynthesis in Fagopyrum tataricum. J. Exp. Botany 2018, 69, 1955–1966. [Google Scholar] [CrossRef] [Green Version]
- Araújo, G.D.S.; Paula-Marinho, S.D.O.; Pinheiro, S.K.D.P.; Miguel, E.D.C.; Lopes, L.D.S.; Marques, E.C.; de Carvalho, H.H.; Gomes-Filho, E. H2O2 priming promotes salt tolerance in maize by protecting chloroplasts ultrastructure and primary metabolites modulation. Plant Sci. 2020, 303, 110774. [Google Scholar] [CrossRef]
- Christensen, S.A.; Santana, E.A.; Alborn, H.T.; Block, A.K.; Chamberlain, C.A. Metabolomics by UHPLC-HRMS reveals the impact of heat stress on pathogen-elicited immunity in maize. Metabolomics 2021, 17, 6. [Google Scholar] [CrossRef]
- Durenne, B.; Blondel, A.; Druart, P.; Fauconnier, M.-L. Epoxiconazole exposure affects terpenoid profiles of oilseed rape plantlets based on a targeted metabolomic approach. Environ. Sci. Pollut. Res. 2019, 26, 17362–17372. [Google Scholar] [CrossRef]
- Poveda, J.; Velasco, P.; de Haro, A.; Johansen, T.; McAlvay, A.; Möllers, C.; Mølmann, J.; Ordiales, E.; Rodríguez, V. Agronomic and Metabolomic Side-Effects of a Divergent Selection for Indol-3-Ylmethylglucosinolate Content in Kale (Brassica oleracea var. acephala). Metabolites 2021, 11, 384. [Google Scholar] [CrossRef] [PubMed]
- Austel, N.; Böttcher, C.; Meiners, T. Chemical defense in Brassicaceae against pollen beetles revealed by metabolomics and flower bud manipulation approaches. Plant Cell Environ. 2021, 44, 519–534. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, Q.; Yu, Y.; Li, X.; Tan, H. Integrated proteomics, metabolomics and physiological analyses for dissecting the toxic effects of halosulfuron-methyl on soybean seedlings (Glycine max merr.). Plant Physiol. Biochem. 2020, 157, 303–315. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Teng, L.; Tang, X.; Long, W.; Wang, Z.; Wu, Y.; Liao, L. Agro-morphological and metabolomics analysis of low nitrogen stress response in Axonopus compressus. AoB Plants 2021, 13, plab022. [Google Scholar] [CrossRef] [PubMed]
- Shaar-Moshe, L.; Hayouka, R.; Roessner, U.; Peleg, Z. Phenotypic and metabolic plasticity shapes life-history strategies under combinations of abiotic stresses. Plant Direct 2019, 3, e00113. [Google Scholar] [CrossRef] [Green Version]
- Abdelrahman, M.; Burritt, D.J.; Tran, L.-S.P. The use of metabolomic quantitative trait locus mapping and osmotic adjustment traits for the improvement of crop yields under environmental stresses. Semin. Cell Dev. Biol. 2017, 83, 86–94. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Lv, J.; Liu, Z.; Liu, Y.; Song, J.; Ma, Y.; Ou, L.; Zhang, X.; Liang, C.; Wang, F.; et al. Integration of Transcriptomics and Metabolomics for Pepper (Capsicum annuum L.) in Response to Heat Stress. Int. J. Mol. Sci. 2019, 20, 5042. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, X.; Zhu, Y.; Zhang, R.; Zhu, Z.; Zhao, T.; Cheng, L.; Gao, L.; Liu, B.; Zhang, X.; Wang, Y. Ionomic and metabolomic analyses reveal the resistance response mechanism to saline-alkali stress in Malus halliana seedlings. Plant Physiol. Biochem. 2019, 147, 77–90. [Google Scholar] [CrossRef]
- Monroy-Velandia, D.; Coy-Barrera, E. Effect of salt stress on growth and metabolite profiles of cape gooseberry (Physalis peruviana L.) along three growth stages. Molecules 2021, 26, 2756. [Google Scholar] [CrossRef]
- Pérez-Díaz, J.; Batista-Silva, W.; Almada, R.; Medeiros, D.B.; Arrivault, S.; Correa, F.; Bastías, A.; Rojas, P.; Beltrán, M.F.; Pozo, M.F.; et al. Prunus Hexokinase 3 genes alter primary C-metabolism and promote drought and salt stress tolerance in Arabidopsis transgenic plants. Sci. Rep. 2021, 11, 7098. [Google Scholar] [CrossRef]
- Stavridou, E.; Voulgari, G.; Michailidis, M.; Kostas, S.; Chronopoulou, E.; Labrou, N.; Madesis, P.; Nianiou-Obeidat, I. Overexpression of A Biotic Stress-Inducible Pvgstu Gene Activates Early Protective Responses in Tobacco under Combined Heat and Drought. Int. J. Mol. Sci. 2021, 22, 2352. [Google Scholar] [CrossRef] [PubMed]
- Reimer, J.J.; Thiele, B.; Biermann, R.T.; Junker-Frohn, L.V.; Wiese-Klinkenberg, A.; Usadel, B.; Wormit, A. Tomato leaves under stress: A comparison of stress response to mild abiotic stress between a cultivated and a wild tomato species. Plant Mol. Biol. 2021, 107, 177–206. [Google Scholar] [CrossRef] [PubMed]
- Larriba, E.; Sánchez-García, A.B.; Martínez-Andújar, C.; Albacete, A.; Pérez-Pérez, J.M. Tissue-Specific Metabolic Reprogramming during Wound-Induced Organ Formation in Tomato Hypocotyl Explants. Int. J. Mol. Sci. 2021, 22, 10112. [Google Scholar] [CrossRef] [PubMed]
- Demirel, U.; Morris, W.L.; Ducreux, L.J.M.; Yavuz, C.; Asim, A.; Tindas, I.; Campbell, R.; Morris, J.A.; Verrall, S.R.; Hedley, P.; et al. Physiological, Biochemical, and Transcriptional Responses to Single and Combined Abiotic Stress in Stress-Tolerant and Stress-Sensitive Potato Genotypes. Front. Plant Sci. 2020, 11, 169. [Google Scholar] [CrossRef] [PubMed]
- Cabral, C.; Wollenweber, B.; António, C.; Rodrigues, A.M.; Ravnskov, S. Aphid infestation in the phyllosphere affects primary metabolic profiles in the arbuscular mycorrhizal hyphosphere. Sci. Rep. 2018, 8, 14442. [Google Scholar] [CrossRef] [Green Version]
- Goddard, M.-L.; Belval, L.; Martin, I.R.; Roth, L.; Laloue, H.; Deglène-Benbrahim, L.; Valat, L.; Bertsch, C.; Chong, J. Arbuscular Mycorrhizal Symbiosis Triggers Major Changes in Primary Metabolism Together With Modification of Defense Responses and Signaling in Both Roots and Leaves of Vitis vinifera. Front. Plant Sci. 2021, 12, 721614. [Google Scholar] [CrossRef]
- Yadav, V.; Arif, N.; Kováč, J.; Singh, V.P.; Tripathi, D.K.; Chauhan, D.K.; Vaculík, M. Structural modifications of plant organs and tissues by metals and metalloids in the environment: A review. Plant Physiol. Biochem. 2020, 159, 100–112. [Google Scholar] [CrossRef]
- Chen, M. The Tea Plant Leaf Cuticle: From Plant Protection to Tea Quality. Front. Plant Sci. 2021, 12, 751547. [Google Scholar] [CrossRef]
- Rastogi, S.; Shah, S.; Kumar, R.; Vashisth, D.; Akhtar, Q.; Kumar, A.; Dwivedi, U.N.; Shasany, A.K. Ocimum metabolomics in response to abiotic stresses: Cold, flood, drought and salinity. PLoS ONE 2019, 14, e0210903. [Google Scholar] [CrossRef]
- Yao, L.; Li, P.; Du, Q.; Quan, M.; Li, L.; Xiao, L.; Song, F.; Lu, W.; Fang, Y.; Zhang, D. Genetic Architecture Underlying the Metabolites of Chlorogenic Acid Biosynthesis in Populus tomentosa. Int. J. Mol. Sci. 2021, 22, 2386. [Google Scholar] [CrossRef]
- Sharma, A.; Wang, J.; Xu, D.; Tao, S.; Chong, S.; Yan, D.; Li, Z.; Yuan, H.; Zheng, B. Melatonin regulates the functional components of photosynthesis, antioxidant system, gene expression, and metabolic pathways to induce drought resistance in grafted Carya cathayensis plants. Sci. Total Environ. 2020, 713, 136675. [Google Scholar] [CrossRef] [PubMed]
- Acevedo, R.M.; Avico, E.H.; González, S.; Salvador, A.R.; Rivarola, M.; Paniego, N.; Nunes-Nesi, A.; Ruiz, O.A.; Sansberro, P.A. Transcript and metabolic adjustments triggered by drought in Ilex paraguariensis leaves. Planta 2019, 250, 445–462. [Google Scholar] [CrossRef] [PubMed]
- Valledor, L.; Guerrero, S.; García-Campa, L.; Meijón, M. Proteometabolomic characterization of apical bud maturation in Pinus pinaster. Tree Physiol. 2020, 41, 508–521. [Google Scholar] [CrossRef] [PubMed]
- Sebastiana, M.; Gargallo-Garriga, A.; Sardans, J.; Pérez-Trujillo, M.; Monteiro, F.; Figueiredo, A.; Maia, M.; Nascimento, R.; Silva, M.S.; Ferreira, A.N.; et al. Metabolomics and transcriptomics to decipher molecular mechanisms underlying ectomycorrhizal root colonization of an oak tree. Sci. Rep. 2021, 11, 8576. [Google Scholar] [CrossRef] [PubMed]
- Chevalier, W.; Moussa, S.-A.; Ottoni, M.M.N.; Dubois-Laurent, C.; Huet, S.; Aubert, C.; Desnoues, E.; Navez, B.; Cottet, V.; Chalot, G.; et al. Multisite evaluation of phenotypic plasticity for specialized metabolites, some involved in carrot quality and disease resistance. PLoS ONE 2021, 16, e0249613. [Google Scholar] [CrossRef]
- Le Gall, H.; Fontaine, J.-X.; Molinié, R.; Pelloux, J.; Mesnard, F.; Gillet, F.; Fliniaux, O. NMR-based Metabolomics to Study the Cold-acclimation Strategy of Two Miscanthus Genotypes. Phytochem. Anal. 2016, 28, 58–67. [Google Scholar] [CrossRef]
- Yin, J.; Gosney, M.J.; Dilkes, B.P.; Mickelbart, M.V. Dark period transcriptomic and metabolic profiling of two diverse Eutrema salsugineum accessions. Plant Direct 2018, 2, e00032. [Google Scholar] [CrossRef] [Green Version]
- Liu, B.; Wang, X.; Li, K.; Cai, Z. Spatially Resolved Metabolomics and Lipidomics Reveal Salinity and Drought-Tolerant Mechanisms of Cottonseeds. J. Agric. Food Chem. 2021, 69, 8028–8037. [Google Scholar] [CrossRef]
- Luo, X.; Li, Z.; Xiao, S.; Ye, Z.; Nie, X.; Zhang, X.; Kong, J.; Zhu, L. Phosphate deficiency enhances cotton resistance to Verticillium dahliae through activating jasmonic acid biosynthesis and phenylpropanoid pathway. Plant Sci. 2020, 302, 110724. [Google Scholar] [CrossRef]
- Panda, A.; Rangani, J.; Parida, A.K. Physiological and metabolic adjustments in the xero-halophyte Haloxylon salicornicum conferring drought tolerance. Physiol. Plant. 2021, 172, 1189–1211. [Google Scholar] [CrossRef]
- Feiner, A.; Pitra, N.; Matthews, P.; Pillen, K.; Wessjohann, L.A.; Riewe, D. Downy mildew resistance is genetically mediated by prophylactic production of phenylpropanoids in hop. Plant Cell Environ. 2020, 44, 323–338. [Google Scholar] [CrossRef] [PubMed]
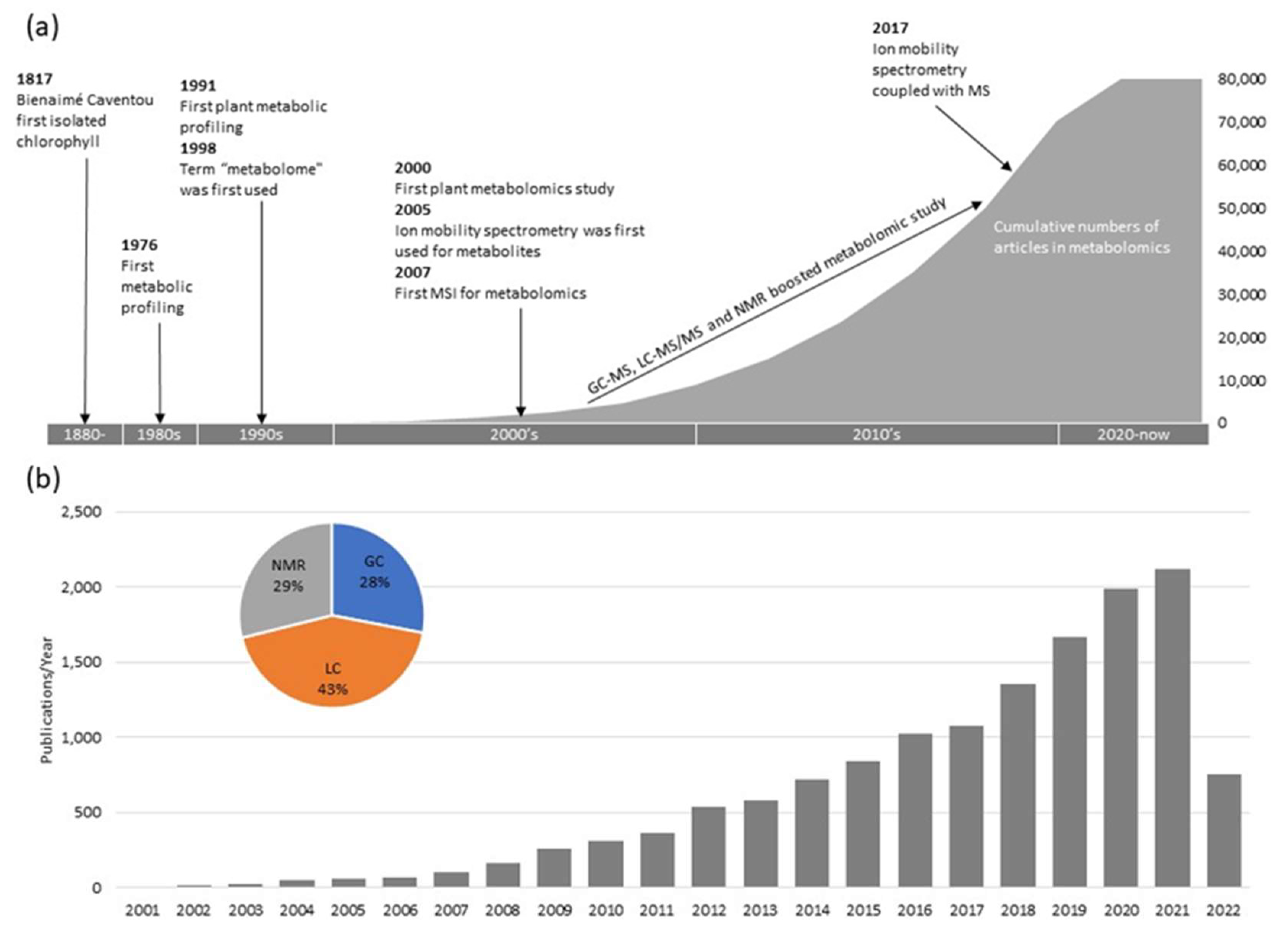
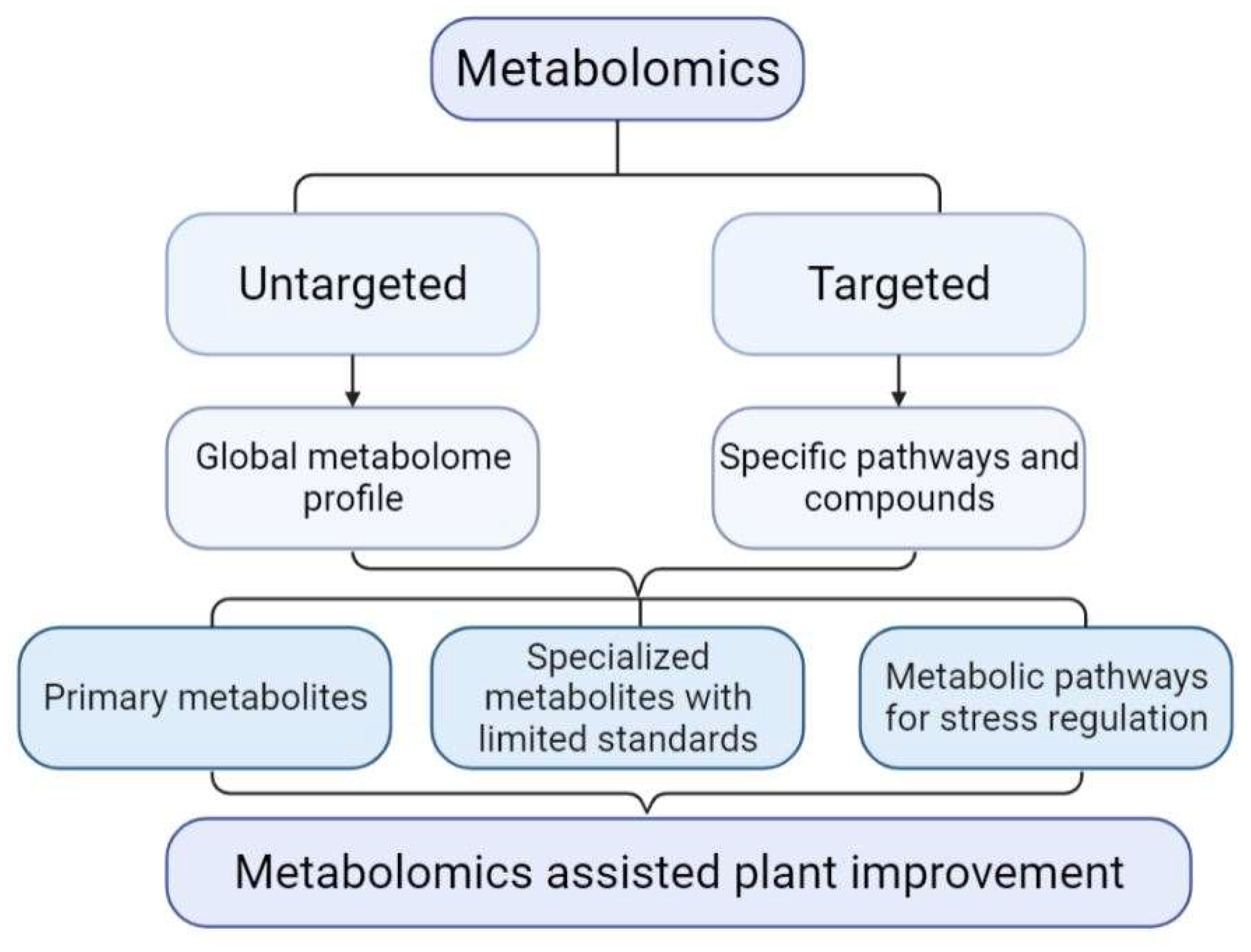
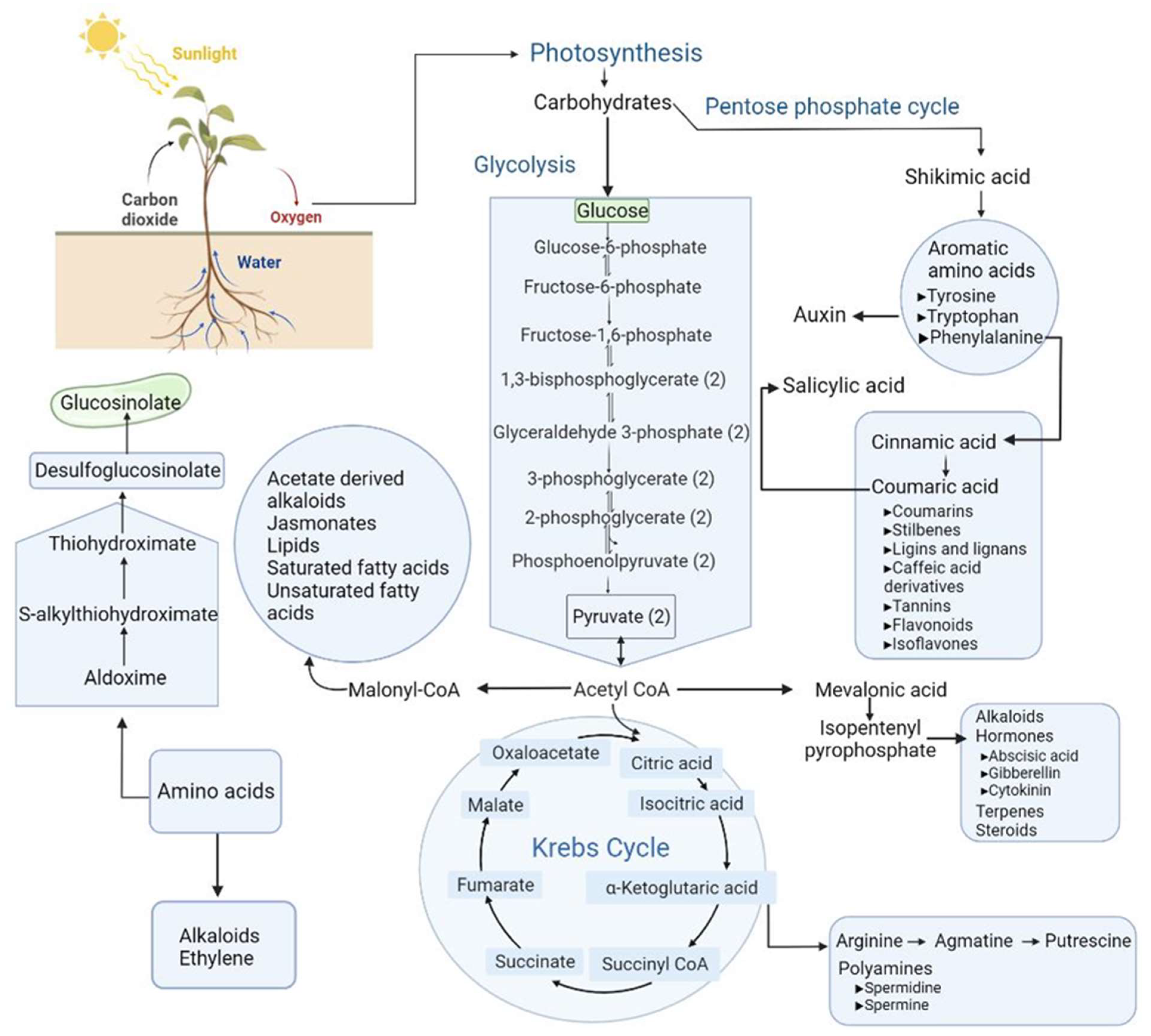
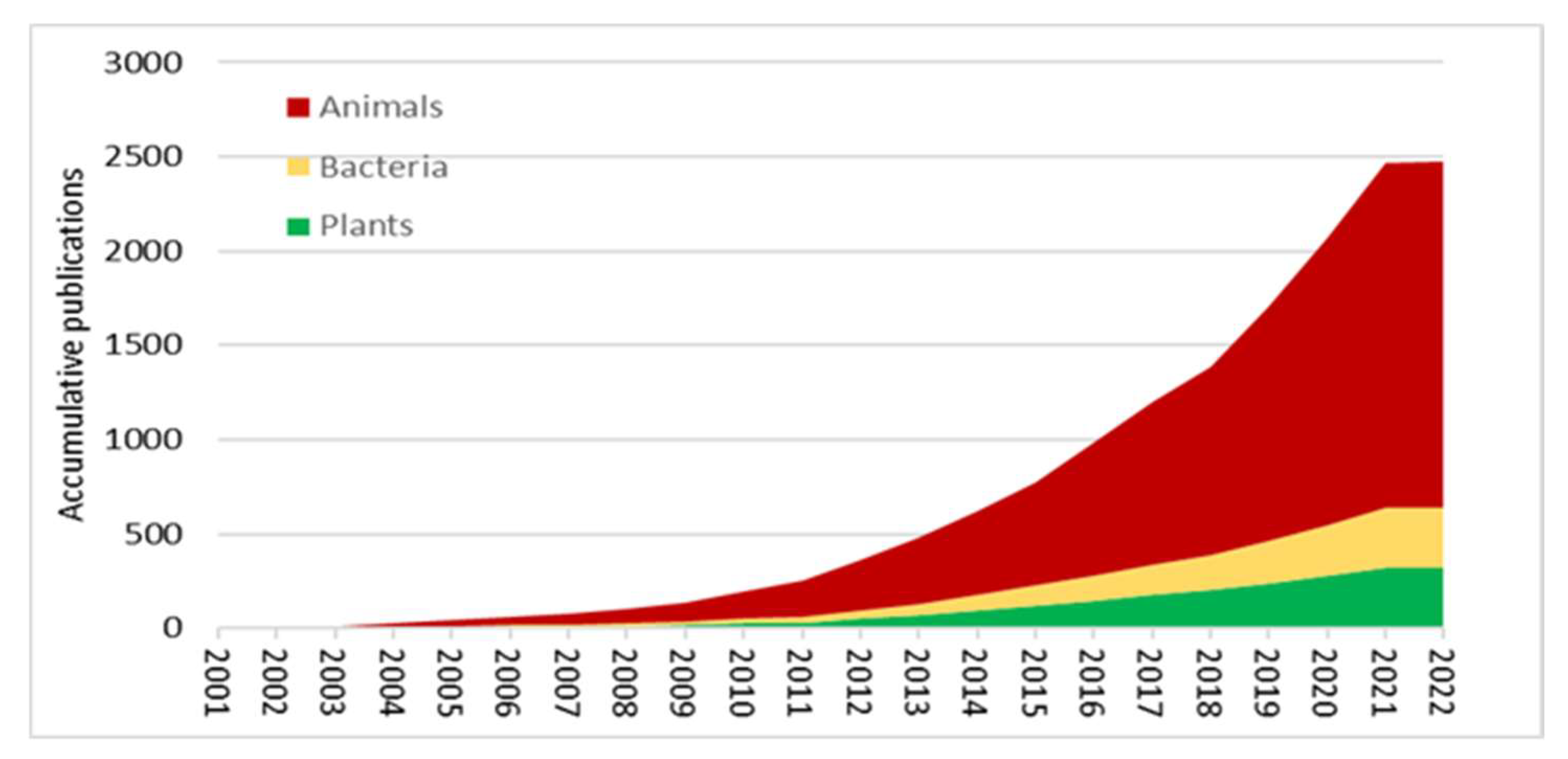
| Plant | Stress | Method | Tissue | Type of Metabolites | Refs |
|---|---|---|---|---|---|
| A. thaliana | Heat shock | GC-MS | Root and shoot | Glyceric acid, maltose, asparagine, glutamine, glycine, and trehalose | [91] |
| LC-MS/MS | Leaf | Phenylalanine-derived metabolites | [92] | ||
| Drought | LC/UPLC-MS/MS; GC-MS | Hormones, MGDG, DGDG, SQDG, PC, PE, PS, PI, and TAG | [93] | ||
| Heat primed, heat shock | UPLC-MS/MS; HILIC/UPLC-MS/MS | Amino acid, carbohydrate, lipid, nucleotide, 2-isopropyl malate, dihydrokaempferol, putrescine, 2-hydroxy laurate, glycerol 3-phosphate, glutathione, ascorbate, tocopherol, and GPC | [94] | ||
| High light, cold | GC-TOF MS; LC-MS/MS | Citrate, gluconic acid, hexose, amino acid, organic acid, sugar | [95] | ||
| GC-TOF-MS | Sucrose phosphate, starch, serine, raffinose, pyruvate, malate, and proline | [96] | |||
| GC-TOF-MS | Sucrose, glucose, and fructose | [97] | |||
| Bacteria | UPLC-MS/MS | Hormone and redox metabolites | [98] | ||
| Genetic modification of AtTSPO | 1H-NMR; HPTLC | Seed and leaf | Fatty acids, lipids, sterols, esters, TAG, FFA, DAG, and starch | [99] | |
| Drought, heat | FT-ICR-MS; GC-TOF-MS | Leaf | Sucrose, sorbitol, coniferyl alcohol, cinnamyl alcohol, and fatty acids | [100] | |
| Brachypodium distachyon | Drought | GC–MS | Leaf | Fatty acid, malic acid, amino acids | [101] |
| Lotus japonicus | Agrobacterium | LC-IonTrap-MS/MS; NMR | Flower buds and petals | Flavonoids, quercetagetin, gossypetin | [102] |
| Physcomitrella patens | Salt, cold, abscisic acid | GC-MS | Protonema tissue | Sugar, amino acid, and organic acid | [103] |
| High night temperatures | GC-TOF-MS | Leaf | Amino acids, sugars, organic acids, phenylpropanoids, phosphates, and polyhydroxy acids | [104] | |
| Oryza sativa | Drought | GC-MS; LC | Leaf | Citric and aconitic acids, benzoic acid, carbohydrates, proline, norvaline, GABA, benzoic acid, TCA cycle acids, and sugars | [105] |
| Salinity | GC-TQMS | Leaf and root | Mannitol, sugar, and organic acid | [106] | |
| Hordeum vulgare | Salinity | LC-MS/MS; MALDI–MSI | Root | PC, fatty acyls, glycerophospholipid, glycerolipid, prenol lipid, polyketide, sphingolipid, DAG, TAG, and SQDAG | [107] |
| GC-MS; LC-MS/MS | Root and leaf | Phytohormones and chlorophyll | [108] | ||
| Drought, bacteria | LC-MS/MS | Leaf | Flavonoid, auxin, flavonol, flavanone, anthocyanin, and hormones | [109] | |
| Medicago sativa; M. truncatula | Drought | LC-TQMS | Leaf and root | Flavonoid, carbohydrate, abscisic acid, and proline | [110] |
| M. truncatula | Drought, Fusarium oxysporium | LC-MS/MS | Organic acid, sugar, citrate, isocitrate, and tetrahydroxychalcone | [111] | |
| Histone deacetylase inhibitor | LC-MS/MS; GC-MS | Seed and seedling | Amino acid, lipid, and carbohydrate; saccharopine, UDP-glucose/UDP-galactose, 1-linolenoylglycerol, 1-linoleoyl-GPI 182, creatine, and N-acetylglutamine | [112] |
| Name | Treatment | Instrument | Cell Type | Metabolites | Refs |
|---|---|---|---|---|---|
| Chlamydomonas reinhardtii | Herbicide | MALDI-MS | Single-cell | Lipids; DGDG, TAG, DGTS | [72] |
| Light/dark | GC-TOF-MS; UPLC | Single-cell | Lipids, nucleic acids, intermediates of glycolysis, TCA metabolites, polyamines | [201] | |
| Euglena gracilis | Light/Dark | GC-MS | Chloroplast | Amino acids, lipid metabolites | [207] |
| Haematococcus pluvialis | Melatonin | LC-MS | Single-cell | Carotenogenic, astaxanthin, and lipids | [208] |
| High light, fulvic acid, and N starvation | LC-MS/MS | Single-cell | Astaxanthin, carbohydrates, lipids | [169] | |
| H. pluviali, Coscinodiscus granii | None * | LDI-HR-MS | Cell wall, single-cell | Photosynthetic pigments | [209] |
| Zygnema sp., | None * | GC-MS | Single-cell | Chlorophylls | [210] |
| Fucus vesiculosus | Various seasons | UPLC-MSN | Single-cell | Chlorophylls, phlorotannin, lipids, and carotenoids | [77] |
| A. thaliana | None * | LMD- LC-MS/MS | Epidermis, cortex, vascular bundles and pith of flowering stem | IAA, JA | [211] |
| None * | LAAPPI-MS/MS | Trichome, single cells, interveinal lamina | Flavonol glycosides, fatty acids, fatty acid esters, galactolipids, and glycosphingolipids | [212] | |
| Hormone, Haloperonospora | NanoLC ESI-MS/MS | Mesophyll, epidermal, and stomatal guard cells | Phytohormones | [200] | |
| Single-walled carbon nanohorn (SWCNH) | GC-MS | Root tip, stem cells | Auxin, serine, methionine, 3,5,7-trihydroxy-4′-methoxy flavone, citraconic acid, hypoxanthine, cellotetraose, 3,4′,5,6,7-methoxyflavone, serotonin, 2,3,4-tri methoxy mandelic acid, epicatechin, furfuryl alcohol, glycolic acid, and ß-sitosterol | [213] | |
| Dark | GC-MS | Leaf mitochondria | Chlorophylls, proline | [214] | |
| Allium cepa, Chlamydomonas reinhardtii | None | LMD-LVC-MS/MS | Epidermis of A. cepa and microalgal cells | [202] | |
| Allium cepa, Fittonia argyroneura | None | LAESI-MS/MS | F. argyroneura leaves, epidermal layers of A. cepa | Acids, carbohydrates, catechol, phthalide, lysine | [215] |
| Catharanthus roseus | None | MSI | Leaf spatial imaging | Terpenoid indole alkaloids | [68] |
| Glycine max | Soil bacteria, Bradyrhizobium japonicum | 21T-FTICR-MS; fLAESI-MS | Root nodule cells | lipids, oligosaccharides, and soyasaponins | [76] |
| Gossypium hirsutum L. | Shading | UPLC-MS/MS | Cotton fibers cells | Amino acids and derivatives, phenylpropanoids, nucleotides and derivatives, lipids, organic acids | [216] |
| Lobelia cardinalis | None | CZE-MS | Hairy root cell cultures | Alkaloids | [205] |
| Oryza sativa | High night temperature | PPESI-MS | Inner endosperm cells | Sugars; malic acid, glutamic acid, ascorbic acid, and Hexose | [203] |
| Picea abies | None | picoPPESI-MS/MS | Parenchymal ray cells, tracheid of the xylem | Organic acids, sugars, most amino acids, glutathione, and abietic acid; coniferin, p-coumaryl alcohol 4-glucoside, and quinic acid | [217] |
| Sonchus arvenis | Dolomite | GC-MS; TLC | Leaf callus, sclerenchyma, parenchyma cells | Pelargonic acid, decanoic acid, and hexadecanoic acid | [206] |
| Vitis vinifera | None | NanoLC-MS/MS | Leaf apoplast | Lipids, phenolic metabolites, and carbohydrates | [218] |
| Zea mays | None | MALDI-MSI; GC-MS | Thylakoid membranes, mesophyll, and bundle sheath cells | 600 metabolites: primary amines, carbonyl groups, carboxylic acids | [219] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Katam, R.; Lin, C.; Grant, K.; Katam, C.S.; Chen, S. Advances in Plant Metabolomics and Its Applications in Stress and Single-Cell Biology. Int. J. Mol. Sci. 2022, 23, 6985. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms23136985
Katam R, Lin C, Grant K, Katam CS, Chen S. Advances in Plant Metabolomics and Its Applications in Stress and Single-Cell Biology. International Journal of Molecular Sciences. 2022; 23(13):6985. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms23136985
Chicago/Turabian StyleKatam, Ramesh, Chuwei Lin, Kirstie Grant, Chaquayla S. Katam, and Sixue Chen. 2022. "Advances in Plant Metabolomics and Its Applications in Stress and Single-Cell Biology" International Journal of Molecular Sciences 23, no. 13: 6985. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms23136985






