Absolute Quantification of Nav1.5 Expression by Targeted Mass Spectrometry
Abstract
:1. Introduction
2. Results
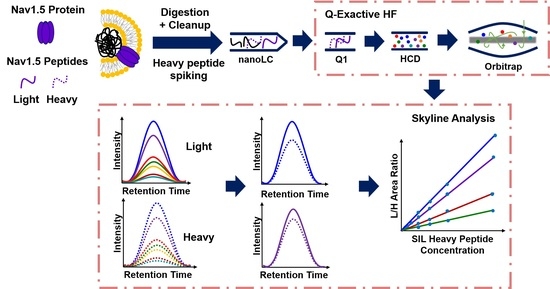
2.1. Trypsin Digestion
2.2. LLOQ of Selected Peptides and Their Corresponding Transitions
2.3. Robustness of the Assay
2.4. Quantitation of SCN5A in Transiently Expressed CHO Cells
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Cell Culture and Harvesting
4.3. Digestion
4.4. LC-MS/MS Analysis
4.5. Data Processing
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dong, C.; Wang, Y.; Ma, A.; Wang, T. Life Cycle of the Cardiac Voltage-Gated Sodium Channel Na. Front. Physiol. 2020, 11, 609733. [Google Scholar] [CrossRef] [PubMed]
- Jiang, D.; Shi, H.; Tonggu, L.; Gamal El-Din, T.M.; Lenaeus, M.J.; Zhao, Y.; Yoshioka, C.; Zheng, N.; Catterall, W.A. Structure of the Cardiac Sodium Channel. Cell 2020, 180, 122–134. [Google Scholar] [CrossRef] [PubMed]
- Abriel, H.; Rougier, J.S.; Jalife, J. Ion channel macromolecular complexes in cardiomyocytes: Roles in sudden cardiac death. Circ. Res. 2015, 116, 1971–1988. [Google Scholar] [CrossRef] [Green Version]
- Han, D.; Tan, H.; Sun, C.; Li, G. Dysfunctional Nav1.5 channels due to SCN5A mutations. Exp. Biol. Med. 2018, 243, 852–863. [Google Scholar] [CrossRef]
- Li, W.; Yin, L.; Shen, C.; Hu, K.; Ge, J.; Sun, A. Variants: Association with Cardiac Disorders. Front. Physiol. 2018, 9, 1372. [Google Scholar] [CrossRef]
- Yang, P.; Koopmann, T.T.; Pfeufer, A.; Jalilzadeh, S.; Schulze-Bahr, E.; Kääb, S.; Wilde, A.A.; Roden, D.M.; Bezzina, C.R. Polymorphisms in the cardiac sodium channel promoter displaying variant in vitro expression activity. Eur. J. Hum. Genet. 2008, 16, 350–357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dumaine, R.; Wang, Q.; Keating, M.T.; Hartmann, H.A.; Schwartz, P.J.; Brown, A.M.; Kirsch, G.E. Multiple mechanisms of Na+ channel–linked long-QT syndrome. Circ. Res. 1996, 78, 916–924. [Google Scholar] [CrossRef]
- Herren, A.W.; Bers, D.M.; Grandi, E. Post-translational modifications of the cardiac Na channel: Contribution of CaMKII-dependent phosphorylation to acquired arrhythmias. Am. J. Physiol. Heart Circ. Physiol. 2013, 305, H431–H445. [Google Scholar] [CrossRef] [Green Version]
- Modell, S.M.; Lehmann, M.H. The long QT syndrome family of cardiac ion channelopathies: A HuGE review. Genet. Med. 2006, 8, 143–155. [Google Scholar] [CrossRef] [Green Version]
- Derangeon, M.; Montnach, J.; Baro, I.; Charpentier, F. Mouse Models of SCN5A-Related Cardiac Arrhythmias. Front. Physiol. 2012, 3, 210. [Google Scholar] [CrossRef] [Green Version]
- Lopez-Santiago, L.F.; Meadows, L.S.; Ernst, S.J.; Chen, C.; Malhotra, J.D.; McEwen, D.P.; Speelman, A.; Noebels, J.L.; Maier, S.K.; Lopatin, A.N.; et al. Sodium channel Scn1b null mice exhibit prolonged QT and RR intervals. J. Mol. Cell. Cardiol. 2007, 43, 636–647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, G.X.; Remme, C.A.; Boukens, B.J.; Belardinelli, L.; Rajamani, S. Overexpression of SCN5A in mouse heart mimics human syndrome of enhanced atrioventricular nodal conduction. Heart Rhythm 2015, 12, 1036–1045. [Google Scholar] [CrossRef]
- Zhang, T.; Yong, S.L.; Tian, X.L.; Wang, Q.K. Cardiac-specific overexpression of SCN5A gene leads to shorter P wave duration and PR interval in transgenic mice. Biochem. Biophys. Res. Commun. 2007, 355, 444–450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brugada, R.; Campuzano, O.; Sarquella-Brugada, G.; Brugada, J.; Brugada, P. Brugada syndrome. Methodist Debakey Cardiovasc. J. 2014, 10, 25–28. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Hernández, M.; Matamoros, M.; Alfayate, S.; Nieto-Marín, P.; Utrilla, R.G.; Tinaquero, D.; de Andrés, R.; Crespo, T.; Ponce-Balbuena, D.; Willis, B.C.; et al. Brugada syndrome trafficking-defective Nav1.5 channels can trap cardiac Kir2.1/2.2 channels. JCI Insight 2018, 3. [Google Scholar] [CrossRef] [Green Version]
- Pfahnl, A.E.; Viswanathan, P.C.; Weiss, R.; Shang, L.L.; Sanyal, S.; Shusterman, V.; Kornblit, C.; London, B.; Dudley, S.C. A sodium channel pore mutation causing Brugada syndrome. Heart Rhythm 2007, 4, 46–53. [Google Scholar] [CrossRef] [Green Version]
- Clerx, M.; Heijman, J.; Collins, P.; Volders, P.G.A. Predicting changes to INa from missense mutations in human SCN5A. Sci. Rep. 2018, 8, 12797. [Google Scholar] [CrossRef]
- Luo, L.; Ning, F.; Du, Y.; Song, B.; Yang, D.; Salvage, S.C.; Wang, Y.; Fraser, J.A.; Zhang, S.; Ma, A.; et al. Calcium-dependent Nedd4-2 upregulation mediates degradation of the cardiac sodium channel Nav1.5: Implications for heart failure. Acta Physiol. 2017, 221, 44–58. [Google Scholar] [CrossRef]
- Remme, C.A.; Bezzina, C.R. Sodium channel (dys)function and cardiac arrhythmias. Cardiovasc. Ther. 2010, 28, 287–294. [Google Scholar] [CrossRef]
- Leoni, A.L.; Gavillet, B.; Rougier, J.S.; Marionneau, C.; Probst, V.; Le Scouarnec, S.; Schott, J.J.; Demolombe, S.; Bruneval, P.; Huang, C.L.; et al. Variable Na(v)1.5 protein expression from the wild-type allele correlates with the penetrance of cardiac conduction disease in the Scn5a(+/−) mouse model. PLoS ONE 2010, 5, e9298. [Google Scholar] [CrossRef] [Green Version]
- Papadatos, G.A.; Wallerstein, P.M.; Head, C.E.; Ratcliff, R.; Brady, P.A.; Benndorf, K.; Saumarez, R.C.; Trezise, A.E.; Huang, C.L.; Vandenberg, J.I.; et al. Slowed conduction and ventricular tachycardia after targeted disruption of the cardiac sodium channel gene Scn5a. Proc. Natl. Acad. Sci. USA 2002, 99, 6210–6215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leong, I.U.; Stuckey, A.; Lai, D.; Skinner, J.R.; Love, D.R. Assessment of the predictive accuracy of five in silico prediction tools, alone or in combination, and two metaservers to classify long QT syndrome gene mutations. BMC Med. Genet. 2015, 16, 34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosati, B.; McKinnon, D. Regulation of ion channel expression. Circ. Res. 2004, 94, 874–883. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Huang, Y.; Li, W.; Wang, Z.; Zhan, S.; Zhou, M.; Yao, Y.; Zeng, Z.; Hou, Y.; Chen, Q.; et al. Post-transcriptional regulation of cardiac sodium channel gene SCN5A expression and function by miR-192-5p. Biochim. Biophys. Acta 2015, 1852, 2024–2034. [Google Scholar] [CrossRef] [Green Version]
- Fahmi, A.I.; Patel, M.; Stevens, E.B.; Fowden, A.L.; John, J.E.; Lee, K.; Pinnock, R.; Morgan, K.; Jackson, A.P.; Vandenberg, J.I. The sodium channel beta-subunit SCN3b modulates the kinetics of SCN5a and is expressed heterogeneously in sheep heart. J. Physiol. 2001, 537, 693–700. [Google Scholar] [CrossRef]
- Yuan, L.; Koivumäki, J.T.; Liang, B.; Lorentzen, L.G.; Tang, C.; Andersen, M.N.; Svendsen, J.H.; Tfelt-Hansen, J.; Maleckar, M.; Schmitt, N.; et al. Investigations of the Navβ1b sodium channel subunit in human ventricle; functional characterization of the H162P Brugada syndrome mutant. Am. J. Physiol. Heart Circ. Physiol. 2014, 306, H1204–H1212. [Google Scholar] [CrossRef] [Green Version]
- Tarradas, A.; Pinsach-Abuin, M.L.; Mackintosh, C.; Llora-Batlle, O.; Perez-Serra, A.; Batlle, M.; Perez-Villa, F.; Zimmer, T.; Garcia-Bassets, I.; Brugada, R.; et al. Transcriptional regulation of the sodium channel gene (SCN5A) by GATA4 in human heart. J. Mol. Cell. Cardiol. 2017, 102, 74–82. [Google Scholar] [CrossRef] [Green Version]
- Scheppach, C.; Robinson, H.P.C. Fluctuation Analysis in Nonstationary Conditions: Single Ca2+ Channel Current in Pyramidal Neurons. Biophys. J. 2017, 113, 2383–2395. [Google Scholar] [CrossRef] [Green Version]
- Verkerk, A.O.; Marchal, G.A.; Zegers, J.G.; Kawasaki, M.; Driessen, A.H.G.; Remme, C.A.; de Groot, J.R.; Wilders, R. Patch-Clamp Recordings of Action Potentials From Human Atrial Myocytes: Optimization Through Dynamic Clamp. Front. Pharmacol. 2021, 12, 649414. [Google Scholar] [CrossRef]
- Laedermann, C.J.; Syam, N.; Pertin, M.; Decosterd, I.; Abriel, H. β1- and β3-voltage-gated sodium channel subunits modulate cell surface expression and glycosylation of Nav1.7 in HEK293 cells. Front. Cell. Neurosci. 2013, 7, 137. [Google Scholar] [CrossRef] [Green Version]
- Lu, P.; Vogel, C.; Wang, R.; Yao, X.; Marcotte, E.M. Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat. Biotechnol. 2007, 25, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Vidova, V.; Spacil, Z. A review on mass spectrometry-based quantitative proteomics: Targeted and data independent acquisition. Anal. Chim. Acta 2017, 964, 7–23. [Google Scholar] [CrossRef] [PubMed]
- Ranade, A.V.; Mukhtarov, R.; An Liu, K.J.; Behrner, M.A.; Sun, B. Characterization of Sample Loss Caused by Competitive Adsorption of Proteins in Vials Using Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis. Langmuir 2019, 35, 4224–4232. [Google Scholar] [CrossRef]
- Sun, B.; Kovatch, J.R.; Badiong, A.; Merbouh, N. Optimization and Modeling of Quadrupole Orbitrap Parameters for Sensitive Analysis toward Single-Cell Proteomics. J. Proteome Res. 2017, 16, 3711–3721. [Google Scholar] [CrossRef] [PubMed]
- Fouda, M.A.; Ruben, P.C. Protein Kinases Mediate Anti-Inflammatory Effects of Cannabidiol and Estradiol Against High Glucose in Cardiac Sodium Channels. Front. Pharmacol. 2021, 12, 668657. [Google Scholar] [CrossRef]
- Salvarani, N.; Crasto, S.; Miragoli, M.; Bertero, A.; Paulis, M.; Kunderfranco, P.; Serio, S.; Forni, A.; Lucarelli, C.; Dal Ferro, M.; et al. The K219T-Lamin mutation induces conduction defects through epigenetic inhibition of SCN5A in human cardiac laminopathy. Nat. Commun. 2019, 10, 2267. [Google Scholar] [CrossRef] [Green Version]
- Westenbroek, R.E.; Bischoff, S.; Fu, Y.; Maier, S.K.; Catterall, W.A.; Scheuer, T. Localization of sodium channel subtypes in mouse ventricular myocytes using quantitative immunocytochemistry. J. Mol. Cell. Cardiol. 2013, 64, 69–78. [Google Scholar] [CrossRef] [Green Version]
- Watanabe, H.; Yang, T.; Stroud, D.M.; Lowe, J.S.; Harris, L.; Atack, T.C.; Wang, D.W.; Hipkens, S.B.; Leake, B.; Hall, L.; et al. Striking In vivo phenotype of a disease-associated human SCN5A mutation producing minimal changes in vitro. Circulation 2011, 124, 1001–1011. [Google Scholar] [CrossRef] [Green Version]
- Yong, S.L.; Ni, Y.; Zhang, T.; Tester, D.J.; Ackerman, M.J.; Wang, Q.K. Characterization of the cardiac sodium channel SCN5A mutation, N1325S, in single murine ventricular myocytes. Biochem. Biophys. Res. Commun. 2007, 352, 378–383. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Dolz-Gaitón, P.; Núñez, M.; Núñez, L.; Barana, A.; Amorós, I.; Matamoros, M.; Pérez-Hernández, M.; de la Fuente, M.G.; Alvarez-López, M.; Macías-Ruiz, R.; et al. Functional characterization of a novel frameshift mutation in the C-terminus of the Nav1.5 channel underlying a Brugada syndrome with variable expression in a Spanish family. PLoS ONE 2013, 8, e81493. [Google Scholar] [CrossRef] [PubMed]
- Ko, S.H.; Lenkowski, P.W.; Lee, H.C.; Mounsey, J.P.; Patel, M.K. Modulation of Na(v)1.5 by beta1- and beta3-subunit co-expression in mammalian cells. Pflugers Arch. 2005, 449, 403–412. [Google Scholar] [CrossRef] [PubMed]
- Plant, L.D.; Xiong, D.; Romero, J.; Dai, H.; Goldstein, S.A.N. Hypoxia Produces Pro-arrhythmic Late Sodium Current in Cardiac Myocytes by SUMOylation of Na. Cell Rep. 2020, 30, 2225–2236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turker, I.; Makiyama, T.; Ueyama, T.; Shimizu, A.; Yamakawa, M.; Chen, P.S.; Vatta, M.; Horie, M.; Ai, T. Telethonin variants found in Brugada syndrome, J-wave pattern ECG, and ARVC reduce peak Na. Pacing Clin. Electrophysiol. 2020, 43, 838–846. [Google Scholar] [CrossRef]
- Trapani, J.G.; Korn, S.J. Control of ion channel expression for patch clamp recordings using an inducible expression system in mammalian cell lines. BMC Neurosci. 2003, 4, 15. [Google Scholar] [CrossRef] [Green Version]
- Fouda, M.A.; Ghovanloo, M.R.; Ruben, P.C. Cannabidiol protects against high glucose-induced oxidative stress and cytotoxicity in cardiac voltage-gated sodium channels. Br. J. Pharmacol. 2020, 177, 2932–2946. [Google Scholar] [CrossRef]
- Sun, B.; Ma, L.; Yan, X.; Lee, D.; Alexander, V.; Hohmann, L.J.; Lorang, C.; Chandrasena, L.; Tian, Q.; Hood, L. N-Glycoproteome of E14.Tg2a mouse embryonic stem cells. PLoS ONE. 2013, 8, e55722. [Google Scholar] [CrossRef]
- Pino, L.K.; Searle, B.C.; Bollinger, J.G.; Nunn, B.; MacLean, B.; MacCoss, M.J. The Skyline ecosystem: Informatics for quantitative mass spectrometry proteomics. Mass Spectrom. Rev. 2020, 39, 229–244. [Google Scholar] [CrossRef] [PubMed]





| Peptide Sequence | MS1 Light M.W. (g/mol) | MS2 Type | MS2 Light M.W. (g/mol) | Neutralized CE * (%) | LLOQ ** Light (fmol) |
|---|---|---|---|---|---|
| HASFLFR | 878.4752 | precursor | 878.4752 | 4 | |
| precursor [M + 1] | 879.4780 | 5 | |||
| precursor [M + 2] | 880.4807 | 7 | |||
| y6 | 740.4090 | 27 | 2 | ||
| y5 | 669.3719 | 1 | |||
| a1 | 110.0713 | 2 | |||
| b2 | 209.1033 | 3 | |||
| VLLEYADK | 951.5266 | precursor | 951.5266 | 0.4 | |
| precursor [M + 1] | 952.5296 | 0.08 | |||
| precursor [M + 2] | 953.5323 | 2 | |||
| y7 | 851.4509 | 22 | 0.2 | ||
| y6 | 738.3668 | 0.2 | |||
| a2 | 625.2828 | 0.3 | |||
| b2 | 213.1598 | 0.4 | |||
| YFFSPTLFR | 1178.6113 | precursor | 1178.6113 | 8 | |
| precursor [M + 1] | 1179.6144 | 0.7 | |||
| precursor [M + 2] | 1180.6172 | 0.3 | |||
| y7 | 867.4723 | 27 | 2 | ||
| y6 | 720.4039 | 0.3 | |||
| y5 | 633.3719 | 0.3 | |||
| b2 | 311.1390 | 0.8 | |||
| YQGFIFDIVTK | 1331.7114 | precursor | 1331.7114 | 5 | |
| precursor [M + 1] | 1332.7145 | 2 × 10 | |||
| precursor [M + 2] | 1333.7173 | 4 | |||
| y9 | 1039.5823 | 27 | 1 | ||
| y6 | 722.4083 | 4 | |||
| a1 | 136.0757 | 6 | |||
| b2 | 292.1292 | 0.1 |
| HASFLFR | VLLEYADK | YFFSPTLFR | YQGFIFDIVTK | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Transition | Slope | R2 **** | Slope | R2 | Slope | R2 | Slope | R2 | |
| M | avg. * | 0.55 | 0.998 | 0.49 | 0.998 | 0.2 | 0.981 | 0.076 | 0.958 |
| stdev. ** | 0.02 | 0.0005 | 0.03 | 0.001 | 0.01 | 0.008 | 0.005 | 0.01 | |
| CV *** | 0.036 | 0.0005 | 0.061 | 0.001 | 0.05 | 0.0082 | 0.066 | 0.01 | |
| M + 1 | avg. | 0.66 | 0.997 | 0.51 | 0.9995 | 0.211 | 0.989 | 0.088 | 0.964 |
| stdev. | 0.08 | 0.002 | 0.04 | 0.0005 | 0.009 | 0.008 | 0.003 | 0.002 | |
| CV | 0.12 | 0.002 | 0.078 | 0.0005 | 0.043 | 0.0081 | 0.034 | 0.0021 | |
| M + 2 | avg. | 0.64 | 0.998 | 0.63 | 0.999 | 0.233 | 0.981 | 0.096 | 0.966 |
| stdev. | 0.07 | 0.0025 | 0.03 | 0.0004 | 0.004 | 0.016 | 0.001 | 0.008 | |
| CV | 0.11 | 0.0025 | 0.048 | 0.0004 | 0.017 | 0.016 | 0.01 | 0.0083 | |
| y9 | avg. | 0.092 | 0.918 | ||||||
| stdev. | N/A | N/A | N/A | 0.05 | 0.1 | ||||
| CV | 0.54 | 0.11 | |||||||
| y7 | avg. | 0.6 | 0.989 | 0.12 | 0.963 | 0.09 | 0.972 | ||
| stdev. | N/A | 0.3 | 0.01 | 0.04 | 0.04 | 0.04 | 0.027 | ||
| CV | 0.5 | 0.01 | 0.33 | 0.042 | 0.44 | 0.028 | |||
| y6 | avg. | 0.6 | 0.989 | 0.58 | 0.996 | 0.20 | 0.957 | 0.2 | 0.8695 |
| stdev. | 0.3 | 0.01 | 0.07 | 0.004 | 0.04 | 0.024 | 0.3 | 0.2 | |
| CV | 0.5 | 0.01 | 0.12 | 0.004 | 0.2 | 0.025 | 1.5 | 0.23 | |
| y5 | avg. | 1.4 | 0.991 | 0.3 | 0.988 | 0.14 | 0.95 | ||
| stdev. | 0.7 | 0.004 | 0.04 | 0.01 | 0.04 | 0.036 | N/A | ||
| CV | 0.5 | 0.004 | 0.13 | 0.01 | 0.29 | 0.038 | |||
| y1 | avg. | 0.8 | 0.993 | ||||||
| stdev. | 0.4 | 0.005 | N/A | N/A | N/A | ||||
| CV | 0.5 | 0.005 | |||||||
| b2 | avg. | 1 | 0.97 | 1 | 0.981 | 0.14 | 0.976 | 0.1 | 0.976 |
| stdev. | 1 | 0.015 | 1 | 0.014 | 0.05 | 0.034 | 0.1 | 0.0044 | |
| CV | 1 | 0.016 | 1 | 0.014 | 0.38 | 0.035 | 1 | 0.0045 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adams, S.L.; Chang, G.; Fouda, M.A.; Kumar, S.; Sun, B. Absolute Quantification of Nav1.5 Expression by Targeted Mass Spectrometry. Int. J. Mol. Sci. 2022, 23, 4177. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms23084177
Adams SL, Chang G, Fouda MA, Kumar S, Sun B. Absolute Quantification of Nav1.5 Expression by Targeted Mass Spectrometry. International Journal of Molecular Sciences. 2022; 23(8):4177. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms23084177
Chicago/Turabian StyleAdams, Sarah L., Ge Chang, Mohamed A. Fouda, Sharwan Kumar, and Bingyun Sun. 2022. "Absolute Quantification of Nav1.5 Expression by Targeted Mass Spectrometry" International Journal of Molecular Sciences 23, no. 8: 4177. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms23084177