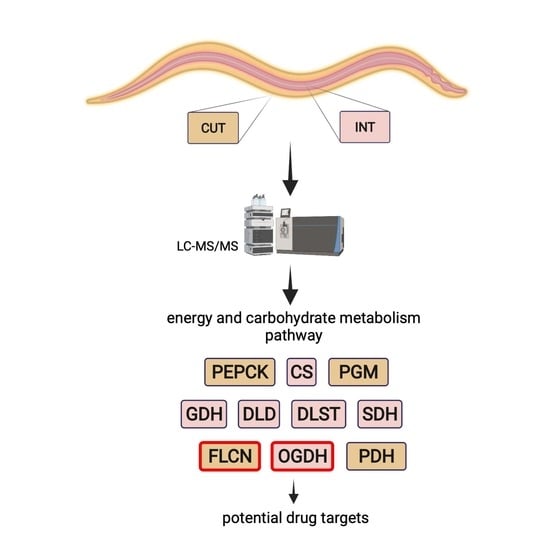
Tandem Mass Tagging (TMT) Reveals Tissue-Specific Proteome of L4 Larvae of Anisakis simplex s. s.: Enzymes of Energy and/or Carbohydrate Metabolism as Potential Drug Targets in Anisakiasis
Abstract
:1. Introduction
2. Results
2.1. Overview of Differentially Regulated Proteins
2.2. Functional Enrichment Analysis and Pathway Identification
2.2.1. Gene Ontology (GO) Analysis
2.2.2. Pathway Identification
2.3. Enzymes Identification
2.4. Tertiary Structure Protein Modeling and Its Detection with Western Blot
2.5. Protein–Protein Interactions (PPIs)
2.5.1. Global PPI Network
2.5.2. Host–Pathogen Protein Interactions
2.6. Allergens Identification
3. Discussion
4. Materials and Methods
4.1. Anisakis Simplex
4.2. Sample Preparation
4.3. Protein Extraction, Preparation, and LC-MS/MS Analysis
4.3.1. Protein Extraction
4.3.2. TMT Labeling and Reversed-Phase Fractionation
4.3.3. LC-MS/MS Analysis and Data Processing
4.3.4. Statistical Analysis
4.4. Results Analysis
4.4.1. Functional Enrichment Analysis and Pathway Identification
4.4.2. Enzymes Identification
4.4.3. 3D Structures Modulation
4.4.4. Western Blot of Selected Proteins
4.4.5. Protein–Protein Interactions Analysis
4.4.6. Allergen Identification
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kelley, L.A.; Sternberg, M.J.E. Protein Structure Prediction on the Web: A Case Study Using the Phyre Server. Nat. Protoc. 2009, 4, 363–371. [Google Scholar] [CrossRef] [Green Version]
- Bouwknegt, M.; Devleesschauwer, B.; Graham, H.; Robertson, L.J.; van der Giessen, J.W. Prioritisation of Food-Borne Parasites in Europe, 2016. Eurosurveillance 2018, 23, 17-00161. [Google Scholar] [CrossRef] [Green Version]
- Stryiński, R.; Łopieńska-Biernat, E.; Carrera, M. Proteomic Insights into the Biology of the Most Important Foodborne Parasites in Europe. Foods 2020, 9, 1403. [Google Scholar] [CrossRef]
- Audicana, M.T.; Kennedy, M.W. Anisakis Simplex: From Obscure Infectious Worm to Inducer of Immune Hypersensitivity. Clin. Microbiol. Rev. 2008, 21, 360–379. [Google Scholar] [CrossRef] [Green Version]
- Aibinu, I.E.; Smooker, P.M.; Lopata, A.L. Anisakis Nematodes in Fish and Shellfish- from Infection to Allergies. Int. J. Parasitol. Parasites Wildl. 2019, 9, 384–393. [Google Scholar] [CrossRef]
- Bao, M.; Pierce, G.J.; Pascual, S.; González-Muñoz, M.; Mattiucci, S.; Mladineo, I.; Cipriani, P.; Bušelić, I.; Strachan, N.J.C. Assessing the Risk of an Emerging Zoonosis of Worldwide Concern: Anisakiasis. Sci. Rep. 2017, 7, 43699. [Google Scholar] [CrossRef] [Green Version]
- Prevalence of Rare Diseases: Bibliographic Data, Orphanet Report Series, Rare Diseases Collection, January 2022, Number 1: Diseases Listed in Alphabetical Order. Available online: https://www.orpha.net/orphacom/cahiers/docs/GB/Prevalence_of_rare_diseases_by_alphabetical_list.pdf (accessed on 24 March 2022).
- EFSA Scientific Opinion on Risk Assessment of Parasites in Fishery Products. EFSA J. 2010, 8, 1543. [CrossRef]
- Baird, F.J.; Gasser, R.B.; Jabbar, A.; Lopata, A.L. Foodborne Anisakiasis and Allergy. Mol. Cell. Probes 2014, 28, 167–174. [Google Scholar] [CrossRef] [Green Version]
- Yorimitsu, N.; Hiraoka, A.; Utsunomiya, H.; Imai, Y.; Tatsukawa, H.; Tazuya, N.; Yamago, H.; Shimizu, Y.; Hidaka, S.; Tanihira, T.; et al. Colonic Intussusception Caused by Anisakiasis: A Case Report and Review of the Literature. Intern. Med. 2013, 52, 223–226. [Google Scholar] [CrossRef] [Green Version]
- Herrador, Z.; Daschner, Á.; Perteguer, M.J.; Benito, A. Epidemiological Scenario of Anisakidosis in Spain Based on Associated Hospitalizations: The Tip of the Iceberg. Clin. Infect. Dis. 2019, 69, 69–76. [Google Scholar] [CrossRef] [Green Version]
- Guardone, L.; Armani, A.; Nucera, D.; Costanzo, F.; Mattiucci, S.; Bruschi, F. Human Anisakiasis in Italy: A Retrospective Epidemiological Study over Two Decades. Parasite 2018, 25, 41. [Google Scholar] [CrossRef] [Green Version]
- Mattiucci, S.; Cipriani, P.; Levsen, A.; Paoletti, M.; Nascetti, G. Molecular Epidemiology of Anisakis and Anisakiasis: An Ecological and Evolutionary Road Map. Adv. Parasitol. 2018, 99, 93–263. [Google Scholar] [CrossRef]
- Mladineo, I.; Popović, M.; Drmić-Hofman, I.; Poljak, V. A Case Report of Anisakis Pegreffii (Nematoda, Anisakidae) Identified from Archival Paraffin Sections of a Croatian Patient. BMC Infect. Dis. 2015, 16, 42. [Google Scholar] [CrossRef] [Green Version]
- Kołodziejczyk, L.; Szostakowska, B.; Sobecka, E.; Szczucki, K.; Stankiewicz, K. First Case of Human Anisakiasis in Poland. Parasitol. Int. 2020, 76, 102073. [Google Scholar] [CrossRef]
- Fiorenza, E.A.; Wendt, C.A.; Dobkowski, K.A.; King, T.L.; Pappaionou, M.; Rabinowitz, P.; Samhouri, J.F.; Wood, C.L. It’s a Wormy World: Meta-analysis Reveals Several Decades of Change in the Global Abundance of the Parasitic Nematodes Anisakis Spp. and Pseudoterranova Spp. in Marine Fishes and Invertebrates. Glob. Change Biol. 2020, 26, 2854–2866. [Google Scholar] [CrossRef]
- Klimpel, S.; Palm, H.W. Anisakid Nematode (Ascaridoidea) Life Cycles and Distribution: Increasing Zoonotic Potential in the Time of Climate Change. In Progress in Parasitology. Parasitology Research Monographs; Mehlhorn, H., Ed.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 201–222. ISBN 978-3-642-21395-3. [Google Scholar]
- Køie, M.; Berland, B.; Burt, M.D.B. Development to Third-Stage Larvae Occurs in the Eggs of Anisakis Simplex and Pseudotetranova Decipiens (Nematoda, Ascaridoidea, Anisakidae). Can. J. Fish. Aquat. Sci. 1995, 52, 134–139. [Google Scholar] [CrossRef]
- Mattiucci, S.; Nascetti, G. Advances and Trends in the Molecular Systematics of Anisakid Nematodes, with Implications for Their Evolutionary Ecology and Host—Parasite Co-Evolutionary Processes. Adv. Parasitol. 2008, 66, 47–148. [Google Scholar]
- Audicana, M.T.; Ansotegui, I.J.; de Corres, L.F.; Kennedy, M.W. Anisakis Simplex: Dangerous—Dead and Alive? Trends Parasitol. 2002, 18, 20–25. [Google Scholar] [CrossRef]
- Kliks, M.M. Anisakiasis in the Western United States: Four New Case Reports from California. Am. J. Trop. Med. Hyg. 1983, 32, 526–532. [Google Scholar] [CrossRef]
- Kagei, N.; Isogaki, H. A Case of Abdominal Syndrome Caused by the Presence of a Large Number of Anisakis Larvae. Int. J. Parasitol. 1992, 22, 251–253. [Google Scholar] [CrossRef]
- Weerasooriya, M.V.; Fujino, T.; Ishii, Y.; Kagei, N. The Value of External Morphology in the Identification of Larval Anisakid Nematodes: A Scanning Electron Microscope Study. Z. Parasitenkd. Parasitol. Res. 1986, 72, 765–778. [Google Scholar] [CrossRef]
- Sohn, W.-M.; Na, B.-K.; Kim, T.H.; Park, T.-J. Anisakiasis: Report of 15 Gastric Cases Caused by Anisakis Type I Larvae and a Brief Review of Korean Anisakiasis Cases. Korean J. Parasitol. 2015, 53, 465–470. [Google Scholar] [CrossRef] [Green Version]
- Kliks, M.M. Human Anisakiasis: An Update. J. Am. Med. Assoc. 1986, 255, 2605. [Google Scholar] [CrossRef]
- Kitaoka, S.; Morielli, A.D.; Zhao, F.Q. FGT-1-Mediated Glucose Uptake Is Defective in Insulin/IGF-like Signaling Mutants in Caenorhabditis Elegans. FEBS Open Bio 2016, 6, 576–585. [Google Scholar] [CrossRef] [Green Version]
- Kitaoka, S.; Morielli, A.D.; Zhao, F.-Q. FGT-1 Is a Mammalian GLUT2-Like Facilitative Glucose Transporter in Caenorhabditis Elegans Whose Malfunction Induces Fat Accumulation in Intestinal Cells. PLoS ONE 2013, 8, e68475. [Google Scholar] [CrossRef]
- Grabda, J. Studies on the Life Cycle and Morphogenesis of Anisakis Simplex (Rudolphi, 1809) (Nematoda: Anisakidae) Cultured in Vitro. Acta Ichthyol. Piscat. 1976, 06, 119–141. [Google Scholar] [CrossRef] [Green Version]
- Gallardo, J.M.; Ortea, I.; Carrera, M. Proteomics and Its Applications for Food Authentication and Food-Technology Research. TrAC—Trends Anal. Chem. 2013, 52, 135–141. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING Database in 2021: Customizable Protein-Protein Networks, and Functional Characterization of User-Uploaded Gene/Measurement Sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Bateman, A.; Martin, M.-J.; Orchard, S.; Magrane, M.; Agivetova, R.; Ahmad, S.; Alpi, E.; Bowler-Barnett, E.H.; Britto, R.; Bursteinas, B.; et al. UniProt: The Universal Protein Knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Cavallero, S.; Lombardo, F.; Su, X.; Salvemini, M.; Cantacessi, C.; D’Amelio, S. Tissue-Specific Transcriptomes of Anisakis Simplex (Sensu Stricto) and Anisakis Pegreffii Reveal Potential Molecular Mechanisms Involved in Pathogenicity. Parasites Vectors 2018, 11, 31. [Google Scholar] [CrossRef] [Green Version]
- Page, A.P.; Stepek, G.; Winter, A.D.; Pertab, D. Enzymology of the Nematode Cuticle: A Potential Drug Target? Int. J. Parasitol. Drugs Drug Resist. 2014, 4, 133–141. [Google Scholar] [CrossRef] [Green Version]
- Basyoni, M.M.A.; Rizk, E.M.A. Nematodes Ultrastructure: Complex Systems and Processes. J. Parasit. Dis. 2016, 40, 1130–1140. [Google Scholar] [CrossRef] [Green Version]
- Sakanari, J.A.; McKerrow, J.H. Identification of the Secreted Neutral Proteases from Anisakis Simplex. J. Parasitol. 1990, 76, 625. [Google Scholar] [CrossRef]
- Matthews, B.E. The Source, Release and Specificity of Proteolytic Enzyme Activity Produced by Anisakis Simplex Larvae (Nematoda: Ascaridida) in Vitro. J. Helminthol. 1984, 58, 175–185. [Google Scholar] [CrossRef]
- Bahlool, Q.Z.M.; Skovgaard, A.; Kania, P.W.; Buchmann, K. Effects of Excretory/Secretory Products from Anisakis Simplex (Nematoda) on Immune Gene Expression in Rainbow Trout (Oncorhynchus Mykiss). Fish Shellfish. Immunol. 2013, 35, 734–739. [Google Scholar] [CrossRef] [Green Version]
- White, R.R.; Artavanis-Tsakonas, K. How Helminths Use Excretory Secretory Fractions to Modulate Dendritic Cells. Virulence 2012, 3, 668–677. [Google Scholar] [CrossRef] [Green Version]
- Laing, R.; Kikuchi, T.; Martinelli, A.; Tsai, I.J.; Beech, R.N.; Redman, E.; Holroyd, N.; Bartley, D.J.; Beasley, H.; Britton, C.; et al. The Genome and Transcriptome of Haemonchus Contortus, a Key Model Parasite for Drug and Vaccine Discovery. Genome Biol. 2013, 14, R88. [Google Scholar] [CrossRef] [Green Version]
- Harder, A. The Biochemistry of Haemonchus Contortus and Other Parasitic Nematodes. In Haemonchus Contortus and Haemonchosis—Past, Present and Future Trends; Gasser, R.B., von Samson-Himmelstjerna, G., Eds.; Elsevier Ltd.: Amsterdam, The Netherlands, 2016; Volume 93, pp. 69–94. [Google Scholar]
- Polak, I.; Łopieńska-Biernat, E.; Stryiński, R.; Mateos, J.; Carrera, M. Comparative Proteomics Analysis of Anisakis Simplex s.s.—Evaluation of the Response of Invasive Larvae to Ivermectin. Genes 2020, 11, 710. [Google Scholar] [CrossRef]
- Mierzejewski, K.; Stryiński, R.; Łopieńska-Biernat, E.; Mateos, J.; Bogacka, I.; Carrera, M. A Complex Proteomic Response of the Parasitic Nematode Anisakis Simplex s.s. to Escherichia Coli Lipopolysaccharide. Mol. Cell. Proteom. 2021, 20, 100166. [Google Scholar] [CrossRef]
- Stryiński, R.; Mateos, J.; Pascual, S.; González, Á.F.; Gallardo, J.M.; Łopieńska-Biernat, E.; Medina, I.; Carrera, M. Proteome Profiling of L3 and L4 Anisakis Simplex Development Stages by TMT-Based Quantitative Proteomics. J. Proteom. 2019, 201, 1–11. [Google Scholar] [CrossRef]
- Carrera, M.; Gallardo, J.M.; Pascual, S.; González, Á.F.; Medina, I. Protein Biomarker Discovery and Fast Monitoring for the Identification and Detection of Anisakids by Parallel Reaction Monitoring (PRM) Mass Spectrometry. J. Proteom. 2016, 142, 130–137. [Google Scholar] [CrossRef] [Green Version]
- Fæste, C.K.; Jonscher, K.R.; Dooper, M.M.W.B.; Egge-Jacobsen, W.; Moen, A.; Daschner, A.; Egaas, E.; Christians, U. Characterisation of Potential Novel Allergens in the Fish Parasite Anisakis Simplex. EuPA Open Proteom. 2014, 4, 140–155. [Google Scholar] [CrossRef] [Green Version]
- Kochanowski, M.; Dąbrowska, J.; Różycki, M.; Karamon, J.; Sroka, J.; Cencek, T. Proteomic Profiling Reveals New Insights into the Allergomes of Anisakis Simplex, Pseudoterranova Decipiens, and Contracaecum Osculatum. J. Parasitol. 2020, 106, 572. [Google Scholar] [CrossRef]
- Kochanowski, M.; Różycki, M.; Dąbrowska, J.; Bełcik, A.; Karamon, J.; Sroka, J.; Cencek, T. Proteomic and Bioinformatic Investigations of Heat-Treated Anisakis Simplex Third-Stage Larvae. Biomolecules 2020, 10, 1066. [Google Scholar] [CrossRef]
- Arcos, S.C.; Ciordia, S.; Roberston, L.; Zapico, I.; Jiménez-Ruiz, Y.; Gonzalez-Muñoz, M.; Moneo, I.; Carballeda-Sangiao, N.; Rodriguez-Mahillo, A.; Albar, J.P.; et al. Proteomic Profiling and Characterization of Differential Allergens in the Nematodes Anisakis Simplex Sensu Stricto and A. Pegreffii. Proteomics 2014, 14, 1547–1568. [Google Scholar] [CrossRef]
- Marzano, V.; Pane, S.; Foglietta, G.; Mortera, S.L.; Vernocchi, P.; Muda, A.O.; Putignani, L. Mass Spectrometry Based-proteomic Analysis of Anisakis Spp.: A Preliminary Study towards a New Diagnostic Tool. Genes 2020, 11, 693. [Google Scholar] [CrossRef]
- Robertson, L.; Arcos, S.C.; Ciordia, S.; Carballeda-sanguiao, N.; Mena, M.D.C.; Sánchez-alonso, I.; Gonzalez-muñoz, M.; Careche, M.; Navas, A. Immunoreactive Proteins in the Esophageal Gland Cells of Anisakis Simplex Sensu Stricto Detected by Maldi-tof/Tof Analysis. Genes 2020, 11, 683. [Google Scholar] [CrossRef]
- Kochanowski, M.; Dąbrowska, J.; Różycki, M.; Sroka, J.; Karamon, J.; Bełcik, A.; Korpysa-Dzirba, W.; Cencek, T. Proteomic Profiling and In Silico Characterization of the Secretome of Anisakis Simplex Sensu Stricto L3 Larvae. Pathogens 2022, 11, 246. [Google Scholar] [CrossRef]
- Plubell, D.L.; Wilmarth, P.A.; Zhao, Y.; Fenton, A.M.; Minnier, J.; Reddy, A.P.; Klimek, J.; Yang, X.; David, L.L.; Pamir, N. Extended Multiplexing of Tandem Mass Tags (TMT) Labeling Reveals Age and High Fat Diet Specific Proteome Changes in Mouse Epididymal Adipose Tissue. Mol. Cell. Proteom. 2017, 16, 873–890. [Google Scholar] [CrossRef] [Green Version]
- Wang, T.; Van Steendam, K.; Dhaenens, M.; Vlaminck, J.; Deforce, D.; Jex, A.R.; Gasser, R.B.; Geldhof, P. Proteomic Analysis of the Excretory-Secretory Products from Larval Stages of Ascaris Suum Reveals High Abundance of Glycosyl Hydrolases. PLoS Negl. Trop. Dis. 2013, 7, e2467. [Google Scholar] [CrossRef] [Green Version]
- Mehrdana, F.; Buchmann, K. Excretory/Secretory Products of Anisakid Nematodes: Biological and Pathological Roles. Acta Vet. Scand. 2017, 59, 42. [Google Scholar] [CrossRef] [Green Version]
- Mladineo, I.; Hrabar, J.; Smodlaka, H.; Palmer, L.; Sakamaki, K.; Keklikoglou, K.; Katharios, P. Functional Ultrastructure of the Excretory Gland Cell in Zoonotic Anisakids (Anisakidae, Nematoda). Cells 2019, 8, 1451. [Google Scholar] [CrossRef] [Green Version]
- Bird, A.F.; Bird, J. The Structure of Nematodes, 2nd ed.; Academic Press: London, UK, 2012; ISBN 032313839X, 9780323138390. [Google Scholar]
- Komuniecki, R.; Tielens, A.G.M. Carbohydrate and Energy Metabolism in Parasitic Helminths. In Molecular Medical Parasitology; Marr, J.J., Nilsen, T.W., Komuniecki, R.W., Eds.; Elsevier: Cambridge, MA, USA, 2003; pp. 339–358. [Google Scholar]
- McKerrow, J.H.; Caffrey, C.; Kelly, B.; Loke, P.; Sajid, M. Proteases in Parasitic Diseases. Annu. Rev. Pathol. Mech. Dis. 2006, 1, 497–536. [Google Scholar] [CrossRef]
- Yang, Y.; Wen, Y.J.; Cai, Y.N.; Vallée, I.; Boireau, P.; Liu, M.Y.; Cheng, S.P. Serine Proteases of Parasitic Helminths. Korean J. Parasitol. 2015, 53, 1–11. [Google Scholar] [CrossRef]
- Molina-Fernández, D.; Benítez, R.; Adroher, F.J.; Malagón, D. Differential Proteolytic Activity in Anisakis Simplex s.s. and Anisakis Pegreffii, Two Sibling Species from the Complex Anisakis Simplex s.l., Major Etiological Agents of Anisakiasis. Acta Trop. 2019, 195, 44–50. [Google Scholar] [CrossRef]
- Timson, D.J. Metabolic Enzymes of Helminth Parasites: Potential as Drug Targets. Curr. Protein Pept. Sci. 2016, 17, 280–295. [Google Scholar] [CrossRef]
- Taylor, C.M.; Wang, Q.; Rosa, B.A.; Huang, S.C.-C.; Powell, K.; Schedl, T.; Pearce, E.J.; Abubucker, S.; Mitreva, M. Discovery of Anthelmintic Drug Targets and Drugs Using Chokepoints in Nematode Metabolic Pathways. PLoS Pathog. 2013, 9, e1003505. [Google Scholar] [CrossRef]
- Hamajima, F.; Nishihara, S.; Yasuhiro, C. Glycolytic and Oxidative Metabolism in the Larvae of a Nematode, Anisakis Sp. Jpn. J. Parasitol. 1969, 18, 196–201. [Google Scholar]
- Chambers, J.W.; Fowler, M.L.; Morris, M.T.; Morris, J.C. The Anti-Trypanosomal Agent Lonidamine Inhibits Trypanosoma Brucei Hexokinase 1. Mol. Biochem. Parasitol. 2008, 158, 202–207. [Google Scholar] [CrossRef]
- Singh, A.R.; Joshi, S.; Arya, R.; Kayastha, A.M.; Srivastava, K.K.; Tripathi, L.M.; Saxena, J.K. Molecular Cloning and Characterization of Brugia Malayi Hexokinase. Parasitol. Int. 2008, 57, 354–361. [Google Scholar] [CrossRef]
- Singh, A.R.; Joshi, S.; Arya, R.; Kayastha, A.M.; Saxena, J.K. Guanidine Hydrochloride and Urea-Induced Unfolding of Brugia Malayi Hexokinase. Eur. Biophys. J. 2010, 39, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Dills, W.L.; Meyer, W.L. Studies on 1-Deoxy-D-Fructose, 1-Deoxy-D-Glucitol, and 1-Deoxy-D-Mannitol as Antimetabolites. Biochemistry 1976, 15, 4506–4512. [Google Scholar] [CrossRef] [PubMed]
- Johnsen, U.; Schönheit, P. Characterization of Cofactor-Dependent and Cofactor-Independent Phosphoglycerate Mutases from Archaea. Extremophiles 2007, 11, 647–657. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Foster, J.M.; Kumar, S.; Fougere, M.; Carlow, C.K.S. Cofactor-Independent Phosphoglycerate Mutase Has an Essential Role in Caenorhabditis Elegans and Is Conserved in Parasitic Nematodes. J. Biol. Chem. 2004, 279, 37185–37190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, O.P.; Vadlamudi, Y.; Liao, Q.; Strodel, B.; Suresh Kumar, M. Molecular Modeling, Dynamics, and an Insight into the Structural Inhibition of Cofactor Independent Phosphoglycerate Mutase Isoform 1 from Wuchereria Bancrofti Using Cheminformatics and Mutational Studies. J. Biomol. Struct. Dyn. 2013, 31, 765–778. [Google Scholar] [CrossRef] [PubMed]
- Crowther, G.J.; Booker, M.L.; He, M.; Li, T.; Raverdy, S.; Novelli, J.F.; He, P.; Dale, N.R.G.; Fife, A.M.; Barker, R.H.; et al. Cofactor-Independent Phosphoglycerate Mutase from Nematodes Has Limited Druggability, as Revealed by Two High-Throughput Screens. PLoS Negl. Trop. Dis. 2014, 8, e2628. [Google Scholar] [CrossRef] [Green Version]
- Berg, J.M.; Tymoczko, J.L.; Gatto Jr, G.J.; Stryer, L. Glycolysis and Gluconeogenesis. In Biochemistry; W. H. Freeman and Company: New York, NY, USA, 2015; pp. 449–494. [Google Scholar]
- Berg, J.M.; Tymoczko, J.L.; Gatto Jr, G.J.; Stryer, L. The Citric Acid Cycle. In Biochemistry; W. H. Freeman and Company: New York, NY, USA, 2015; pp. 495–522. [Google Scholar]
- Iglesias, L.; Malagón, D.; Valero, A.; Benítez, R.; Javier Adroher, F. CO2-Fixing Enzymes during Moulting from Third Larval to Fourth Larval Stage of Anisakis Simplex and Hysterothylacium Aduncum (Nematoda: Anisakidae). Parasitol. Res. 2005, 96, 212–215. [Google Scholar] [CrossRef]
- Dávila, C.; Malagón, D.; Valero, A.; Benítez, R.; Adroher, F.J. Anisakis Simplex: CO2-Fixing Enzymes and Development throughout the in Vitro Cultivation from Third Larval Stage to Adult. Exp. Parasitol. 2006, 114, 10–15. [Google Scholar] [CrossRef]
- Prichard, R.K. Regulation of Pyruvate Kinase and Phosphoenolpyruvate Carboxykinase Activity in Adult Fasciola Hepatica (Trematoda). Int. J. Parasitol. 1976, 6, 227–233. [Google Scholar] [CrossRef]
- Tielens, A.G.M.; van den Heuvel, J.M.; van den Bergh, S.G. Differences in Intermediary Energy Metabolism between Juvenile and Adult Fasciola Hepatica. Mol. Biochem. Parasitol. 1987, 24, 273–281. [Google Scholar] [CrossRef] [Green Version]
- Köhler, P. Metabolic Role of Pyruvate Kinase in the Trematode Dicrocoelium Dendriticum. Comp. Biochem. Physiol. Part B: Comp. Biochem. 1974, 49, 335–344. [Google Scholar] [CrossRef]
- Behm, C.A.; Bryant, C. Regulatory Properties of a Partially Purified Preparation of Pyruvate Kinase from Fasciola Hepatica. Int. J. Parasitol. 1980, 10, 107–114. [Google Scholar] [CrossRef]
- Harmych, S.; Arnette, R.; Komuniecki, R. Role of Dihydrolipoyl Dehydrogenase (E3) and a Novel E3-Binding Protein in the NADH Sensitivity of the Pyruvate Dehydrogenase Complex from Anaerobic Mitochondria of the Parasitic Nematode, Ascaris Suum. Mol. Biochem. Parasitol. 2002, 125, 135–146. [Google Scholar] [CrossRef]
- Huang, Y.-J.; Walker, D.; Chen, W.; Klingbeil, M.; Komuniecki, R. Expression of Pyruvate Dehydrogenase Isoforms during the Aerobic/Anaerobic Transition in the Development of the Parasitic NematodeAscaris Suum:Altered Stoichiometry of Phosphorylation/Inactivation. Arch. Biochem. Biophys. 1998, 352, 263–270. [Google Scholar] [CrossRef]
- Huang, Y.-J.; Komuniecki, R. Cloning and Characterization of a Putative Testis-Specific Pyruvate Dehydrogenase β Subunit from the Parasitic Nematode, Ascaris Suum1Note: Nucleotide Sequence Data Reported in This Paper Is Available in the GenBankTM Data Base under the Accession Number AF013755.1. Mol. Biochem. Parasitol. 1997, 90, 391–394. [Google Scholar] [CrossRef]
- Komuniecki, R.; Rhee, R.; Bhat, D.; Duran, E.; Sidawy, E.; Song, H. The Pyruvate Dehydrogenase Complex from the Parasitic Nematode Ascaris Suum: Novel Subunit Composition and Domain Structure of the Dihydrolipoyl Transacetylase Component. Arch. Biochem. Biophys. 1992, 296, 115–121. [Google Scholar] [CrossRef]
- Thissen, J.; Komuniecki, R. Phosphorylation and Inactivation of the Pyruvate Dehydrogenase from the Anaerobic Parasitic Nematode, Ascaris Suum. Stoichiometry and Amino Acid Sequence around the Phosphorylation Sites. J. Biol. Chem. 1988, 263, 19092–19097. [Google Scholar] [CrossRef]
- Rahman, M.M.; Rosu, S.; Joseph-Strauss, D.; Cohen-Fix, O. Down-Regulation of Tricarboxylic Acid (TCA) Cycle Genes Blocks Progression through the First Mitotic Division in Caenorhabditis Elegans Embryos. Proc. Natl. Acad. Sci. USA 2014, 111, 2602–2607. [Google Scholar] [CrossRef] [Green Version]
- Diaz, F.; Komuniecki, R. Characterization of the α-Ketoglutarate Dehydrogenase Complex from Fasciola Hepatica: Potential Implications for the Role of Calcium in the Regulation of Helminth Mitochondrial Metabolism. Mol. Biochem. Parasitol. 1996, 81, 243–246. [Google Scholar] [CrossRef]
- Pawełczyk, T.; Angielski, S. Cooperation of Ca2+ and PH in Regulation of the Activity of the 2-Oxoglutarate Dehydrogenase Complex and Its Components from Bovine Kidney Cortex. Acta Biochim. Pol. 1984, 31, 289–305. [Google Scholar]
- Zhang, X.; Tomar, N.; Kandel, S.M.; Audi, S.H.; Cowley, A.W.; Dash, R.K. Substrate- and Calcium-Dependent Differential Regulation of Mitochondrial Oxidative Phosphorylation and Energy Production in the Heart and Kidney. Cells 2021, 11, 131. [Google Scholar] [CrossRef] [PubMed]
- Chin, R.M.; Fu, X.; Pai, M.Y.; Vergnes, L.; Hwang, H.; Deng, G.; Diep, S.; Lomenick, B.; Meli, V.S.; Monsalve, G.C.; et al. The Metabolite α-Ketoglutarate Extends Lifespan by Inhibiting ATP Synthase and TOR. Nature 2014, 510, 397–401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woodford, M.R.; Baker-Williams, A.J.; Sager, R.A.; Backe, S.J.; Blanden, A.R.; Hashmi, F.; Kancherla, P.; Gori, A.; Loiselle, D.R.; Castelli, M.; et al. The Tumor Suppressor Folliculin Inhibits Lactate Dehydrogenase A and Regulates the Warburg Effect. Nat. Struct. Mol. Biol. 2021, 28, 662–670. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H. Studies on Lactic Dehydrogenase Activity in Parasitic Helminths. Korean J. Parasitol. 1967, 5, 5. [Google Scholar] [CrossRef] [PubMed]
- Barrett, J. Biochemistry of Parasitic Helminths; Macmillan Education: London, UK, 1981; ISBN 978-0-333-25669-5. [Google Scholar]
- vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg Effect: The Metabolic Requirements of Cell Proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ehsan, M.; Wang, W.; Gadahi, J.A.; Hasan, M.W.; Lu, M.; Wang, Y.; Liu, X.; Haseeb, M.; Yan, R.; Xu, L.; et al. The Serine/Threonine-Protein Phosphatase 1 From Haemonchus Contortus Is Actively Involved in Suppressive Regulatory Roles on Immune Functions of Goat Peripheral Blood Mononuclear Cells. Front. Immunol. 2018, 9, 1627. [Google Scholar] [CrossRef] [Green Version]
- Palevich, N.; Maclean, P.H.; Subbaraj, A.K.; Cao, M. A Multimodal Metabolomics Approach to Elucidate the Trigger Components of Rumen Fluid for Larval Exsheathment in a Model Gastrointestinal Parasitic Nematode, Haemonchus Contortus. bioRxiv 2021. [Google Scholar] [CrossRef]
- Łopieńska-Biernat, E.; Paukszto, Ł.; Jastrzębski, J.P.; Makowczenko, K.; Stryiński, R. Genes Expression and in Silico Studies of Functions of Trehalases, a Highly Dispersed Anisakis Simplex s. l. Specific Gene Family. Int. J. Biol. Macromol. 2019, 129, 957–964. [Google Scholar] [CrossRef]
- Łopieńska-Biernat, E.; Paukszto, Ł.; Jastrzębski, J.P.; Myszczyński, K.; Polak, I.; Stryiński, R. Genome-Wide Analysis of Anisakis Simplex Sensu Lato: The Role of Carbohydrate Metabolism Genes in the Parasite’s Development. Int. J. Parasitol. 2019, 49, 933–943. [Google Scholar] [CrossRef]
- Braeckman, B.P. Intermediary Metabolism. In WormBook; The C. elegans Research Community, WormBook. doi/10.1895/wormbook.1.146.1. Available online: http://www.wormbook.org (accessed on 24 March 2022).
- Watts, J.L.; Ristow, M. Lipid and Carbohydrate Metabolism in Caenorhabditis Elegans. Genetics 2017, 207, 413–446. [Google Scholar] [CrossRef]
- Yilmaz, L.S.; Li, X.; Nanda, S.; Fox, B.; Schroeder, F.; Walhout, A.J. Modeling Tissue-relevant Caenorhabditis Elegans Metabolism at Network, Pathway, Reaction, and Metabolite Levels. Mol. Syst. Biol. 2020, 16, e9649. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Chai, Q.-Y.; Liu, C.H. The Ubiquitin System: A Critical Regulator of Innate Immunity and Pathogen–Host Interactions. Cell. Mol. Immunol. 2016, 13, 560–576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harada, S.; Inaoka, D.K.; Ohmori, J.; Kita, K. Diversity of Parasite Complex II. Biochim. Biophys. Acta (BBA)—Bioenerg. 2013, 1827, 658–667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Hellemond, J.J.; Klei, A. van der; Weelden, SusanneW.H. van; Tielens, A.G.M. Biochemical and Evolutionary Aspects of Anaerobically Functioning Mitochondria. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2003, 358, 205–215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Hellemond, J.J.; Klockiewicz, M.; Gaasenbeek, C.P.H.; Roos, M.H.; Tielens, A.G.M. Rhodoquinone and Complex II of the Electron Transport Chain in Anaerobically Functioning Eukaryotes. J. Biol. Chem. 1995, 270, 31065–31070. [Google Scholar] [CrossRef] [Green Version]
- Moore, H.W.; Folkers, K. Coenzyme Q. LXII. Structure and Synthesis of Rhodoquinone, a Natural Aminoquinone of the Coenzyme Q Group 1. J. Am. Chem. Soc. 1965, 87, 1409–1410. [Google Scholar] [CrossRef]
- Otero, L.; Martínez-Rosales, C.; Barrera, E.; Pantano, S.; Salinas, G. Complex I and II Subunit Gene Duplications Provide Increased Fitness to Worms. Front. Genet. 2019, 10, 1043. [Google Scholar] [CrossRef]
- Takamiya, S.; Kita, K.; Wang, H.; Weinstein, P.P.; Hiraishi, A.; Oya, H.; Aoki, T. Developmental Changes in the Respiratory Chain of Ascaris Mitochondria. Biochim. Biophys. Acta (BBA)—Bioenerg. 1993, 1141, 65–74. [Google Scholar] [CrossRef]
- Fioravanti, C.F.; Walker, D.J.; Sandhu, P.S. Metabolic Transition in the Development of Hymenolepis Diminuta (Cestoda). Parasitol. Res. 1998, 84, 777–782. [Google Scholar] [CrossRef]
- Matsumoto, J.; Sakamoto, K.; Shinjyo, N.; Kido, Y.; Yamamoto, N.; Yagi, K.; Miyoshi, H.; Nonaka, N.; Katakura, K.; Kita, K.; et al. Anaerobic NADH-Fumarate Reductase System Is Predominant in the Respiratory Chain of Echinococcus Multilocularis, Providing a Novel Target for the Chemotherapy of Alveolar Echinococcosis. Antimicrob. Agents Chemother. 2008, 52, 164–170. [Google Scholar] [CrossRef] [Green Version]
- Xu, Q.; Zhao, Z.; Liang, P.; Wang, S.; Li, F.; Jin, S.; Zhang, J. Identification of Novel Nematode Succinate Dehydrogenase Inhibitors: Virtual Screening Based on Ligand-pocket Interactions. Chem. Biol. Drug Des. 2022. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, R.; Srivastava, A.K. Biochemical Composition and Metabolic Pathways of Filarial Worms Setaria Cervi: Search for New Antifilarial Agents. J. Helminthol. 2007, 81, 261–280. [Google Scholar] [CrossRef] [PubMed]
- PARASITE. Risk Assessment with Integrated Tools in EU Fish Production Value Chains. Available online: http://parasite-project.eu/project (accessed on 17 February 2022).
- Levsen, A.; Svanevik, C.S.; Cipriani, P.; Mattiucci, S.; Gay, M.; Hastie, L.C.; Bušelić, I.; Mladineo, I.; Karl, H.; Ostermeyer, U.; et al. A Survey of Zoonotic Nematodes of Commercial Key Fish Species from Major European Fishing Grounds—Introducing the FP7 PARASITE Exposure Assessment Study. Fish. Res. 2018, 202, 4–21. [Google Scholar] [CrossRef]
- Stryiński, R.; Mateos, J.; Łopieńska-Biernat, E.; Carrera, M. Shotgun Proteomics for L3 and L4 Anisakis Simplex Development Stages. In Shotgun Proteomics: Methods and Protocols, Methods in Molecular Biology; Carrera, M., Mateos, J., Eds.; Springer: New York, NY, USA, 2021; pp. 59–75. ISBN 978-1-0716-1177-7. [Google Scholar]
- Carrera, M.; Cañas, B.; López-Ferrer, D.; Piñeiro, C.; Vázquez, J.; Gallardo, J.M. Fast Monitoring of Species-Specific Peptide Biomarkers Using High-Intensity-Focused-Ultrasound-Assisted Tryptic Digestion and Selected MS/MS Ion Monitoring. Anal. Chem. 2011, 83, 5688–5695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niu, M.; Cho, J.-H.; Kodali, K.; Pagala, V.; High, A.A.; Wang, H.; Wu, Z.; Li, Y.; Bi, W.; Zhang, H.; et al. Extensive Peptide Fractionation and y 1 Ion-Based Interference Detection Method for Enabling Accurate Quantification by Isobaric Labeling and Mass Spectrometry. Anal. Chem. 2017, 89, 2956–2963. [Google Scholar] [CrossRef] [Green Version]
- Käll, L.; Canterbury, J.D.; Weston, J.; Noble, W.S.; MacCoss, M.J. Semi-Supervised Learning for Peptide Identification from Shotgun Proteomics Datasets. Nat. Methods 2007, 4, 923–925. [Google Scholar] [CrossRef]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. G:Profiler: A Web Server for Functional Enrichment Analysis and Conversions of Gene Lists (2019 Update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef] [Green Version]
- Bu, D.; Luo, H.; Huo, P.; Wang, Z.; Zhang, S.; He, Z.; Wu, Y.; Zhao, L.; Liu, J.; Guo, J.; et al. KOBAS-i: Intelligent Prioritization and Exploratory Visualization of Biological Functions for Gene Enrichment Analysis. Nucleic Acids Res. 2021, 49, W317–W325. [Google Scholar] [CrossRef]
- Gotz, S.; Garcia-Gomez, J.M.; Terol, J.; Williams, T.D.; Nagaraj, S.H.; Nueda, M.J.; Robles, M.; Talon, M.; Dopazo, J.; Conesa, A. High-Throughput Functional Annotation and Data Mining with the Blast2GO Suite. Nucleic Acids Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [Green Version]
- Cheng, J.; Baldi, P. A Machine Learning Information Retrieval Approach to Protein Fold Recognition. Bioinformatics 2006, 22, 1456–1463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y. I-TASSER Server for Protein 3D Structure Prediction. BMC Bioinform. 2008, 9, 40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bates, P.A.; Kelley, L.A.; MacCallum, R.M.; Sternberg, M.J.E. Enhancement of Protein Modeling by Human Intervention in Applying the Automatic Programs 3D-JIGSAW and 3D-PSSM. Proteins Struct. Funct. Genet. 2001, 45, 39–46. [Google Scholar] [CrossRef]
- Vallat, B.K.; Pillardy, J.; Májek, P.; Meller, J.; Blom, T.; Cao, B.; Elber, R. Building and Assessing Atomic Models of Proteins from Structural Templates: Learning and Benchmarks. Proteins Struct. Funct. Bioinform. 2009, 76, 930–945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benkert, P.; Künzli, M.; Schwede, T. QMEAN Server for Protein Model Quality Estimation. Nucleic Acids Res. 2009, 37, W510–W514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arnold, K.; Bordoli, L.; Rgen Kopp, J.; Schwede, T. The SWISS-MODEL Workspace: A Web-Based Environment for Protein Structure Homology Modelling. Bioinformatics 2006, 22, 195–201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berjanskii, M.; Zhou, J.; Liang, Y.; Lin, G.; Wishart, D.S. Resolution-by-Proxy: A Simple Measure for Assessing and Comparing the Overall Quality of NMR Protein Structures. J. Biomol. NMR 2012, 53, 167–180. [Google Scholar] [CrossRef]
- Mcguffin, L.J.; Buenavista, M.T.; Roche, D.B. The ModFOLD4 Server for the Quality Assessment of 3D Protein Models. Nucleic Acids Res. 2013, 41, W368–W372. [Google Scholar] [CrossRef]
- Smolinska, N.; Kiezun, M.; Dobrzyn, K.; Rytelewska, E.; Kisielewska, K.; Gudelska, M.; Zaobidna, E.; Bogus-Nowakowska, K.; Wyrebek, J.; Bors, K.; et al. Expression of Chemerin and Its Receptors in the Porcine Hypothalamus and Plasma Chemerin Levels during the Oestrous Cycle and Early Pregnancy. Int. J. Mol. Sci. 2019, 20, 3887. [Google Scholar] [CrossRef] [Green Version]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING V10: Protein-Protein Interaction Networks, Integrated over the Tree of Life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef]
- Doncheva, N.T.; Morris, J.H.; Gorodkin, J.; Jensen, L.J. Cytoscape StringApp: Network Analysis and Visualization of Proteomics Data. J. Proteome Res. 2019, 18, 623–632. [Google Scholar] [CrossRef] [PubMed]
- Ammari, M.G.; Gresham, C.R.; McCarthy, F.M.; Nanduri, B. HPIDB 2.0: A Curated Database for Host–Pathogen Interactions. Database 2016, 2016, 103. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Nanduri, B. HPIDB—A Unified Resource for Host-Pathogen Interactions. BMC Bioinform. 2010, 11, S16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maurer-Stroh, S.; Krutz, N.L.; Kern, P.S.; Gunalan, V.; Nguyen, M.N.; Limviphuvadh, V.; Eisenhaber, F.; Gerberick, G.F. AllerCatPro—Prediction of Protein Allergenicity Potential from the Protein Sequence. Bioinformatics 2019, 35, 3020–3027. [Google Scholar] [CrossRef] [PubMed] [Green Version]









| Term Category * | Term Name | Term ID | Adjusted p-Value | Term Size | Input Data Size |
|---|---|---|---|---|---|
| INTESTINE | |||||
| MF | serine hydrolase activity | GO:0017171 | 1.495 × 10−9 | 115 | 15 |
| MF | serine-type peptidase activity | GO:0008236 | 1.495 × 10−9 | 115 | 15 |
| MF | peptidase activity | GO:0008233 | 1.038 × 10−6 | 417 | 22 |
| MF | oxidoreductase activity | GO:0016491 | 3.416 × 10−4 | 488 | 20 |
| MF | calcium-dependent phospholipid binding | GO:0005544 | 4.901 × 10−4 | 19 | 5 |
| BP | tricarboxylic acid cycle | GO: 0006099 | 3.291 × 10−7 | 18 | 7 |
| BP | cellular respiration | GO:0045333 | 1.083 × 10−6 | 32 | 8 |
| BP | energy derivation by oxidation of organic compounds | GO:0015980 | 1.338 × 10−6 | 43 | 8 |
| BP | aerobic respiration | GO:0009060 | 1.446 × 10−5 | 29 | 7 |
| BP | proteolysis | GO:0006508 | 1.596 × 10−4 | 598 | 23 |
| CC | mitochondrial membrane | GO:0031966 | 6.598 × 10−6 | 64 | 8 |
| CC | mitochondrial envelope | GO:0005740 | 1.882 × 10−5 | 73 | 8 |
| CC | mitochondrion | GO:0005739 | 1.402 × 10−4 | 165 | 10 |
| CC | organelle envelope | GO:0031967 | 1.713 × 10−4 | 97 | 8 |
| CC | envelope | GO:0031975 | 1.713 × 10−4 | 97 | 8 |
| CUTICLE | |||||
| MF | structural constituent of cuticle | GO:0042302 | 6.155 × 10−15 | 103 | 16 |
| MF | structural molecule activity | GO:0005198 | 1.010 × 10−7 | 296 | 16 |
| MF | hexokinase activity | GO:0004396 | 1.472 × 10−3 | 6 | 3 |
| MF | glucose binding | GO:0005536 | 1.472 × 10−3 | 6 | 3 |
| MF | glutathione hydrolase activity | GO:0036374 | 2.562 × 10−3 | 7 | 3 |
| BP | cellular glucose homeostasis | GO:0001678 | 8.682 × 10−4 | 6 | 3 |
| BP | nucleobase-containing small molecule metabolic process | GO:0055086 | 9.613 × 10−4 | 173 | 8 |
| BP | glucose homeostasis | GO:0042593 | 1.513 × 10−3 | 7 | 3 |
| BP | carbohydrate homeostasis | GO:0033500 | 1.513 × 10−3 | 7 | 3 |
| BP | small molecule metabolic process | GO:0044281 | 2.182 × 10−3 | 497 | 12 |
| CC | extracellular region | GO:0005576 | 3.644 × 10−2 | 130 | 5 |
| CC | extracellular space | GO:0005615 | 1.324 × 10−2 | 56 | 4 |
| Anisakis Tissue-Proteome | AllerCatPro Prediction | ||||
|---|---|---|---|---|---|
| Tissue | UniProt Accession | Protein Name | UniProt/NCBI Accession | Protein Name | Organism |
| Intestine | A0A3P6Q2Z8 | Uncharacterized protein | L7UZ85 | Der f 24 | Dermatophagoides farinae |
| Intestine | A0A0M3JS03 | 60S acidic ribosomal protein P2 | Q9UUZ6 | Asp f 8 | Aspergillus fumigatus |
| Intestine | A0A0M3K232 | Voltage-dependent anion-selective channel protein 3 | Q1HR57 | Aed a 6 | Aedes aegypti |
| Intestine | A0A0M3JYW9 | Fructose-bisphosphate aldolase | B5DGM7 | Sal s 3 | Salmo salar |
| Intestine | A0A0M3KBB5 | Gelsolin-like protein 1 | Q8MVU3 | Der f 16 | Dermatophagoides farinae |
| Cuticle | A0A0M3K208 | Uncharacterized protein | G1FMP3 | Ani s 24 kD | Anisakis simplex |
| Cuticle | A0A3P6N545 | Uncharacterized protein | P59704 | Aspartyl protease inhibitor | Trichostrongylus colubriformis |
| Cuticle | A0A158PPI4 | Uncharacterized protein | E1BI98 | Bos d Collagen 140 kD | Bos taurus |
| Cuticle | A0A0M3JZM7 | Col_cuticle_N domain-containing protein | P02465 | Bos d alpha2I | Bos taurus |
| Cuticle | A0A0M3JZS8 | Uncharacterized protein | O93484 | Onc m alpha2I | Oncorhynchus mykiss |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stryiński, R.; Mateos, J.; Carrera, M.; Jastrzębski, J.P.; Bogacka, I.; Łopieńska-Biernat, E. Tandem Mass Tagging (TMT) Reveals Tissue-Specific Proteome of L4 Larvae of Anisakis simplex s. s.: Enzymes of Energy and/or Carbohydrate Metabolism as Potential Drug Targets in Anisakiasis. Int. J. Mol. Sci. 2022, 23, 4336. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms23084336
Stryiński R, Mateos J, Carrera M, Jastrzębski JP, Bogacka I, Łopieńska-Biernat E. Tandem Mass Tagging (TMT) Reveals Tissue-Specific Proteome of L4 Larvae of Anisakis simplex s. s.: Enzymes of Energy and/or Carbohydrate Metabolism as Potential Drug Targets in Anisakiasis. International Journal of Molecular Sciences. 2022; 23(8):4336. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms23084336
Chicago/Turabian StyleStryiński, Robert, Jesús Mateos, Mónica Carrera, Jan Paweł Jastrzębski, Iwona Bogacka, and Elżbieta Łopieńska-Biernat. 2022. "Tandem Mass Tagging (TMT) Reveals Tissue-Specific Proteome of L4 Larvae of Anisakis simplex s. s.: Enzymes of Energy and/or Carbohydrate Metabolism as Potential Drug Targets in Anisakiasis" International Journal of Molecular Sciences 23, no. 8: 4336. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms23084336