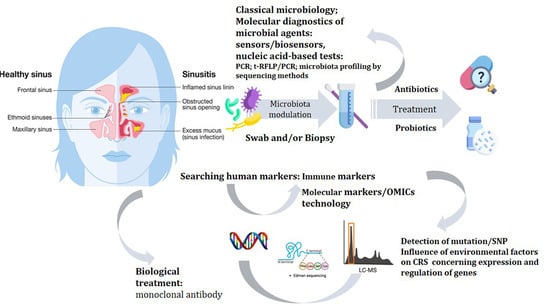
Chronic Rhinosinusitis—Microbiological Etiology, Potential Genetic Markers, and Diagnosis
Abstract
:1. Introduction—Sinus Function, Immunology of Healthy Sinuses
2. Changes That Occur in the Sinuses of Humans with Acute and Chronic Rhinosinusitis
3. The Natural Microbiota of the Sinuses
4. Etiological Agents of Sinusitis
4.1. CRS as a Social Disease
4.2. Changing Sinus Microbiota in Chronic Conditions—Current State of Knowledge
5. From Classical Microbiology to Molecular and OMICS Methods
5.1. Contemporary Research Trends in CRS Diagnostics
5.1.1. Sensors
5.1.2. Nucleic Acid-Based Tests
Microbiota Detection
Human Factors and Susceptibility to CRF
- CRS and polymorphism genes in relation to inflammation reaction and innate immunity
- The human leukocyte antigen (HLA)
- Genes responsible for tissue remodeling
- Genes encoding xenobiotic-metabolizing enzymes
- Taste receptors
- Association between mutation in the CFTR gene and CRS
- Genes in arachidonic acid metabolism associated with CRS
Nucleic Acid-Based Test Problems
5.1.3. Influence of Environmental Factors on CRS concerning Expression and Regulation of Genes
5.1.4. OMICs Technologies
6. The Problem in the Treatment of CRS—Antibiotic Resistance
7. Treatment with Probiotics
8. Biological Treatment
9. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jones, N. The nose and paranasal sinuses physiology and anatomy. Adv. Drug Deliv. Rev. 2001, 51, 5–19. [Google Scholar] [CrossRef] [PubMed]
- Cappello, Z.J.; Minutello, K.; Dublin, A.B. Anatomy, Head and Neck, Nose Paranasal Sinuses. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2023. [Google Scholar]
- Lane, A.P. The Role of Innate Immunity in the Pathogenesis of Chronic Rhinosinusitis. Curr. Allergy Asthma Rep. 2009, 9, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Vickery, T.W.; Ramakrishnan, V.R. Bacterial Pathogens and The Microbiome. Otolaryngol. Clin. N. Am. 2017, 50, 29–47. [Google Scholar] [CrossRef] [PubMed]
- Tai, J.; Han, M.S.; Kwak, J.; Kim, T.H. Association between microbiota and nasal mucosal diseases in terms of immunity. Int. J. Mol. Sci. 2021, 22, 4744. [Google Scholar] [CrossRef]
- Workman, A.D.; Kohanski, M.A.; Cohen, N.A. Biomarkers in Chronic Rhinosinusitis with Nasal Polyps. Immunol. Allergy Clin. N. Am. 2018, 38, 679–692. [Google Scholar] [CrossRef]
- Nam, K.-Y.; Kim, J.-B. Treatment of dental implant-related maxillary sinusitis with functional endoscopic sinus surgery in combination with an intra-oral approach. J. Korean Assoc. Oral Maxillofac. Surg. 2014, 40, 87–90. [Google Scholar] [CrossRef]
- Dykewicz, M.S.; Hamilos, D.L. Rhinitis and sinusitis. J. Allergy Clin. Immunol. 2010, 125 (Suppl. S2), S103–S115. [Google Scholar] [CrossRef]
- Michalik, M.; Podbielska-Kubera, A.; Basińska, A.M.; Szewc, M.; Gałęcka, M.; Schwiertz, A. Alteration of indicator gut microbiota in patients with chronic sinusitis. Immun. Inflamm. Dis. 2023, 11, e996. [Google Scholar] [CrossRef] [PubMed]
- Marcus, S.; Roland, L.T.; DelGaudio, J.M.; Wise, S.K. The relationship between allergy and chronic rhinosinusitis. Laryngoscope Investig. Otolaryngol. 2019, 4, 13–17. [Google Scholar] [CrossRef]
- Li, S.; Zhao, C.-J.; Hua, H.-L.; Deng, Y.-Q.; Tao, Z.-Z. The association between allergy and sinusitis: A cross-sectional study based on NHANES 2005–2006. Allergy Asthma Clin. Immunol. 2021, 17, 135. [Google Scholar] [CrossRef] [PubMed]
- Fokkens, W.J.; Lund, V.J.; Hopkins, C.; Hellings, P.W.; Kern, R.; Reitsma, S.; Toppila-Salmi, S.; Bernal-Sprekelsen, M.; Mullol, J.; Alobid, I.; et al. European PositionPaper on Rhinosinusitis and Nasal Polyps 2020. Rhinology 2020, 50, 1–464. [Google Scholar] [CrossRef]
- Grayson, J.W.; Hopkins, C.; Mori, E.; Senior, B.; Harvey, R.J. Contemporary Classification of Chronic Rhinosinusitis Beyond Polyps vs No Polyps: A Review. JAMA Otolaryngol. Head Neck Surg. 2020, 146, 831–838, Erratum in: JAMA Otolaryngol. Head Neck Surg. 2020, 146, 876. [Google Scholar] [CrossRef] [PubMed]
- Harrass, S.; Yi, C.; Chen, H. Chronic Rhinosinusitis and Alzheimer’s Disease—A Possible Role for the Nasal Microbiome in Causing Neurodegeneration in the Elderly. Int. J. Mol. Sci. 2021, 22, 11207. [Google Scholar] [CrossRef]
- Sivasubramaniam, R.; Douglas, R. The microbiome and chronic rhinosinusitis. World J. Otorhinolaryngol.—Head Neck Surg. 2018, 4, 216–221. [Google Scholar] [CrossRef] [PubMed]
- Mahdavinia, M.; Keshavarzian, A.; Tobin, M.C.; Landay, A.L.; Schleimer, R.P. A comprehensive review of the nasal microbiome in chronic rhinosinusitis (CRS). Clin. Exp. Allergy 2016, 46, 21–41. [Google Scholar] [CrossRef]
- Cope, E.K.; Lynch, S.V. Novel microbiome-based therapeutics for chronic rhinosinusitis. Curr. Allergy Asthma Rep. 2015, 15, 504. [Google Scholar] [CrossRef]
- Ramakrishnan, V.R.; Hauser, L.J.; Frank, D.N. The sinonasal bacterial microbiome in health and disease. Curr. Opin. Otolaryngol. Head Neck Surg. 2016, 24, 20–25. [Google Scholar] [CrossRef] [PubMed]
- Lam, K.; Schleimer, R.; Kern, R.C. The Etiology and Pathogenesis of Chronic Rhinosinusitis: A Review of Current Hypotheses. Curr. Allergy Asthma Rep. 2015, 15, 41. [Google Scholar] [CrossRef]
- Ferguson, M. Rhinosinusitis in oral medicine and dentistry. Aust. Dent. J. 2014, 59, 289–295. [Google Scholar] [CrossRef]
- Stevens, W.W.; Lee, R.J.; Schleimer, R.P.; Cohen, N.A. Chronic rhinosinusitis pathogenesis. J. Allergy Clin. Immunol. 2015, 136, 1442–1453. [Google Scholar] [CrossRef]
- Manes, R.P.; Batra, P.S. Etiology, diagnosis and management of chronic rhinosinusitis. Expert Rev. Anti-Infect. Ther. 2013, 11, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Cho, D.-Y.; Hunter, R.C.; Ramakrishnan, V.R. Ramakrishnan VR. The microbiome and chronic rhinosinusitis. Immunol. Allergy Clin. N. Am. 2020, 40, 251–263. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.W.; Kim, D.Y.; Choi, S.; Won, S.; Kang, H.R.; Yi, H. Microbiome profiling of uncinate tissue and nasal polyps in patients with chronic rhinosinusitis using swab and tissue biopsy. PLoS ONE 2021, 16, e0249688. [Google Scholar] [CrossRef] [PubMed]
- Cervin, A.U. The Potential for Topical Probiotic Treatment of Chronic Rhinosinusitis, a Personal Perspective. Front. Cell. Infect. Microbiol. 2018, 7, 530. [Google Scholar] [CrossRef]
- Park, I.H.; Lee, J.S.; Park, J.H.; Kang, S.H.; Hong, S.M.; Park, I.S.; Yoon, J.H.; Hong, S.J. Comparison of the human microbiome in adults and children with chronic rhinosinusitis. PLoS ONE 2020, 15, e0242770. [Google Scholar] [CrossRef] [PubMed]
- Park, S.C.; Park, I.-H.; Lee, J.S.; Park, S.M.; Kang, S.H.; Hong, S.-M.; Byun, S.-H.; Jung, Y.G.; Hong, S.J. Microbiome of Unilateral Chronic Rhinosinusitis: A Controlled Paired Analysis. Int. J. Environ. Res. Public Health 2021, 18, 9878. [Google Scholar] [CrossRef]
- Lee, J.T.; Frank, D.N.; Ramakrishnan, V. Microbiome of the paranasal sinuses: Update and literature review. Am. J. Rhinol. Allergy 2016, 30, 3–16. [Google Scholar] [CrossRef]
- Psaltis, A.J.; Mackenzie, B.W.; Cope, E.K.; Ramakrishnan, V.R. Unraveling the role of the microbiome in chronic rhinosinusitis. J. Allergy Clin. Immunol. 2022, 149, 1513–1521. [Google Scholar] [CrossRef]
- Krawczyk, B.; Michalik, M.; Fordon, M.; Wysocka, M.; Samet, A.; Nowicki, B. Escherichia coli Strains with Virulent Factors Typical for Uropathogens were Isolated from Sinuses from Patients with Chronic Rhinosinusitis—Case Report. Pathogens 2020, 9, 318. [Google Scholar] [CrossRef]
- Michalik, M.; Samet, A.; Marszałek, A.; Krawczyk, B.; Kotłowski, R.; Nowicki, A.; Anyszek, T.; Nowicki, S.; Kur, J.; Nowicki, B. Intra-operative biopsy in chronic sinusitis detects pathogenic Escherichia coli that carry fimG/H, fyuA and agn43 genes coding biofilm formation. PLoS ONE 2018, 13, e0192899. [Google Scholar] [CrossRef]
- Szyfter, W.; Kruk-Zagajewska, A.; Bartochowska, A.; Borucki, Ł. Intracranial complications from sinusitis. Otolaryngol. Pol. 2015, 69, 6–11. [Google Scholar] [CrossRef]
- Teo, S.M.; Mok, D.; Pham, K.; Kusel, M.; Serralha, M.; Troy, N.; Holt, B.J.; Hales, B.J.; Walker, M.L.; Hollams, E.; et al. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe 2015, 17, 704–715. [Google Scholar] [CrossRef]
- Michalik, M. The innovative laryngological cannula facilitates the precise collection of highly diagnostic material from the nose and sinuses in order to improve the diagnostic and therapeutic process. New Med. 2020, 24, 130–134. [Google Scholar] [CrossRef]
- Zhu, M.; Gao, X.; Zhu, Z.; Hu, X.; Zhou, H.; Liu, J. The roles of nasal nitric oxide in diagnosis and endotypes of chronic rhinosinusitis with nasal polyps. J. Otolaryngol.—Head Neck Surg. 2020, 49, 68. [Google Scholar] [CrossRef] [PubMed]
- Thaler, E.R.; Lee, D.D.; Hanson, C.W. Diagnosis of rhinosinusitis with a colorimetric sensor array. J. Breath Res. 2008, 2, 037016. [Google Scholar] [CrossRef] [PubMed]
- Broza, Y.Y.; Braverman, I.; Haick, H. Breath volatolomics for diagnosing chronic rhinosinusitis. Int. J. Nanomed. 2018, 13, 4661–4670. [Google Scholar] [CrossRef]
- Strohsahl, C.M.; Miller, B.L.; Krauss, T.D. Detection of methicillin-resistant Staphylococcus aureus (MRSA) using the NanoLantern Biosensor. In Frontiers in Pathogen Detection: From Nanosensors to Systems; International Society for Optics and Photonics: Bellingham, DC, USA, 2009; Volume 7167, pp. 193–205. [Google Scholar] [CrossRef]
- Das, A.; Kumar, P.; Swain, S. Recent advances in biosensor based endotoxin detection. Biosens. Bioelectron. 2014, 51, 62–75. [Google Scholar] [CrossRef] [PubMed]
- Bassiouni, A.; Cleland, E.J.; Psaltis, A.; Vreugde, S.; Wormald, P.-J. Sinonasal microbiome sampling: A comparison of techniques. PLoS ONE 2015, 10, e0123216. [Google Scholar] [CrossRef]
- Blackwood, C.B.; Marsh, T.; Kim, S.-H.; Paul, E.A. Terminal restriction fragment length polymorphism data analysis for quantitative comparison of microbial communities. Appl. Environ. Microbiol. 2003, 69, 926–932. [Google Scholar] [CrossRef] [PubMed]
- Stressmann, F.A.; Rogers, G.B.; Chan, S.W.; Howarth, P.H.; Harries, P.G.; Bruce, K.D.; Salib, R.J. Characterization of bacterial community diversity in chronic rhinosinusitis infections using novel culture-independent techniques. Am. J. Rhinol. Allergy 2011, 25, e133–e140. [Google Scholar] [CrossRef]
- Bharti, R.; Grimm, D.G. Current challenges and best-practice protocols for microbiome analysis. Brief. Bioinform. 2021, 22, 178–193. [Google Scholar] [CrossRef] [PubMed]
- Hauser, L.J.; Feazel, L.M.; Ir, D.; Fang, R.; Wagner, B.D.; Robertson, C.E.; Frank, D.N.; Ramakrishnan, V.R.; Hauser, L.J.; Feazel, L.M.; et al. Sinus culture poorly predicts resident microbiota. Int. Forum Allergy Rhinol. 2015, 5, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Biswas, K.; Mackenzie, B.W.; Waldvogel-Thurlow, S.; Middleditch, M.; Jullig, M.; Zoing, M.; Taylor, M.W.; Douglas, R.G. Differentially Regulated Host Proteins Associated with Chronic Rhinosinusitis Are Correlated with the Sinonasal Microbiome. Front. Cell. Infect. Microbiol. 2017, 7, 504. [Google Scholar] [CrossRef] [PubMed]
- Stevens, W.W.; Peters, A.T.; Tan, B.K.; Klingler, A.I.; Poposki, J.A.; Hulse, K.E.; Grammer, L.C.; Welch, K.C.; Smith, S.S.; Conley, D.B.; et al. Associations between Inflammatory Endotypes and Clinical Presentations in Chronic Rhinosinusitis. J. Allergy Clin. Immunol. Pract. 2019, 7, 2812–2820.e3. [Google Scholar] [CrossRef] [PubMed]
- Kato, A.; Schleimer, R.P.; Bleier, B.S. Mechanisms and pathogenesis of chronic rhinosinusitis. J. Allergy Clin. Immunol. 2022, 149, 1491–1503. [Google Scholar] [CrossRef] [PubMed]
- Yoo, F.; Suh, J.D. What is the evidence for genetics in chronic rhinosinusitis? Curr. Opin. Otolaryngol. Head Neck Surg. 2017, 25, 54–63. [Google Scholar] [CrossRef]
- Buysschaert, I.D.; Grulois, V.; Eloy, P.; Jorissen, M.; Rombaux, P.; Bertrand, B.; Collet, S.; Bobic, S.; Vlaminck, S.; Hellings, P.W.; et al. Genetic evidence for a role of IL33 in nasal polyposis. Allergy 2010, 65, 616–622. [Google Scholar] [CrossRef]
- Castano, R.; Bossé, Y.; Endam, L.M.; Desrosiers, M. Evidence of association of interleukin-1 receptor-like 1 gene polymorphisms with chronic rhinosinusitis. Am. J. Rhinol. Allergy 2009, 23, 377–384. [Google Scholar] [CrossRef]
- Endam, L.M.; Bossé, Y.; Filali-Mouhim, A.; Cormier, C.; Boisvert, P.; Boulet, L.; Hudson, T.J.; Desrosiers, M. Polymorphisms in the interleukin-22 receptor alpha-1 gene are associated with severe chronic rhinosinusitis. Otolaryngol. Neck Surg. 2009, 140, 741–747. [Google Scholar] [CrossRef]
- Henmyr, V.; Lind-Halldén, C.; Halldén, C.; Säll, T.; Carlberg, D.; Bachert, C.; Cardell, L.-O. Chronic rhinosinusitis patients show accumulation of genetic variants in PARS2. PLoS ONE 2016, 11, e0158202. [Google Scholar] [CrossRef]
- Platt, M.; Metson, R.; Stankovic, K. Gene-expression signatures of nasal polyps associated with chronic rhinosinusitis and aspirin-sensitive asthma. Curr. Opin. Allergy Clin. Immunol. 2009, 9, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Akyigit, A.; Keles, E.; Etem, E.O.; Ozercan, I.; Akyol, H.; Sakallioglu, O.; Karlidag, T.; Polat, C.; Kaygusuz, I.; Yalcin, S. Genetic polymorphism of antioxidant enzymes in eosinophilic and non-eosinophilic nasal polyposis. Eur. Arch. Oto-Rhino-Laryngol. 2017, 274, 267–273. [Google Scholar] [CrossRef] [PubMed]
- Keles, B.; Cora, T.; Acar, H.; Arbag, H.; Inan, Z.; Ozturk, K.; Ozer, B. Evaluation of HLA-A, -B, -Cw, and -DRB1 alleles frequency in Turkish patients with nasal polyposis. Otolaryngol. Head Neck Surg. 2008, 139, 580–585. [Google Scholar] [CrossRef] [PubMed]
- Luxenberger, W.; Posch, U.; Berghold, A.; Hofmann, T.; Lang-Loidolt, D. HLA patterns in patients with nasal polyposis. Eur. Arch. Oto-Rhino-Laryngol. 2000, 257, 137–139. [Google Scholar] [CrossRef]
- Salomon, B.L.; Leclerc, M.; Tosello, J.; Ronin, E.; Piaggio, E.; Cohen, J.L. Tumor Necrosis Factor α and Regulatory T Cells in Oncoimmunology. Front. Immunol. 2018, 9, 444. [Google Scholar] [CrossRef]
- Tomassen, P.; Vandeplas, G.; Van Zele, T.; Cardell, L.-O.; Arebro, J.; Olze, H.; Förster-Ruhrmann, U.; Kowalski, M.L.; Olszewska-Ziąber, A.; Holtappels, G.; et al. Inflammatory endotypes of chronic rhinosinusitis based on cluster analysis of biomarkers. J. Allergy Clin. Immunol. 2016, 137, 1449–1456.e4. [Google Scholar] [CrossRef]
- Berghea, E.C.; Popa, O.M.; Meirosu, M.; Popa, L.O.; Bara, C.; Bumbacea, R.S. Association of TNF-alpha gene polymorphism with nasal polyposis in Romanian asthmatic patients. Rom. J. Rhinol. 2014, 4, 149–155. [Google Scholar]
- Erbek, S.S.; Yurtcu, E.; Erbek, S.; Atac, F.B.; Sahin, F.I.; Cakmak, O. Proinflammatory cytokine single nucleotide polymorphisms in nasal polyposis. Arch. Otolaryngol. Head Neck Surg. 2007, 133, 705–709. [Google Scholar] [CrossRef] [PubMed]
- Szabó, K.; Kiricsi, Á.; Révész, M.; Vóna, I.; Szabó, Z.; Bella, Z.; Polyánka, H.; Kadocsa, E.; Kemény, L.; Széll, M.; et al. The -308 G>A SNP of TNFA is a factor predisposing to chronic rhinosinusitis associated with nasal polyposis in aspirin-sensitive Hungarian individuals: Conclusions of a genetic study with multiple stratifications. Int. Immunol. 2013, 25, 383–388. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, S.; Xu, J.; Zhang, R.; Zhu, H.; Wu, Y.; Zhu, L.; Li, J.; Chen, L. Etanercept as a new therapeutic option for cytokine release syndrome following chimeric antigen receptor T cell therapy. Exp. Hematol. Oncol. 2021, 10, 16. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.S.; Wilson, J.A.; Bennett, M.; Pearson, J.P. Mucin gene expression in nasal polyps. Acta Oto-Laryngol. 2005, 125, 618–624. [Google Scholar] [CrossRef]
- Liu, L.; Yan, C.; Tao, S. Association of MUC2, MUC5AC and MUC5B genes with the recurrence of nasal polyps. Exp. Ther. Med. 2020, 20, 1808–1814. [Google Scholar] [CrossRef]
- Levchenko, A.S.; Piskunov, V.S.; Konoplya, N.A.; Bushueva, O.Y.; Raspopov, A.A.; Mezentseva, O.Y.; Polonikov, A.V. Genetic Aspects of Chronic Rhinosinusitis. Russ. J. Genet. 2018, 54, 910–918. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, H.; Lin, P.; Zhang, G. Glutathione S-transferase gene polymorphisms and risk of nasal or colorectal polyposis. Biosci. Rep. 2019, 39, BSR20181226. [Google Scholar] [CrossRef] [PubMed]
- Özcan, C.; Tamer, L.; Ates, N.A.; Görür, K. The glutathione-S-transferase gene polymorphisms (Gstt1, Gstm1, and Gstp1) in patients with non-allergic nasal polyposis. Eur. Arch. Oto-Rhino-Laryngol. 2010, 267, 227–232. [Google Scholar] [CrossRef]
- Cătană, I.V.; Popp, R.A.; Ioan, V.P.; Cătană, A.; Rădeanu, D.; Maniu, A.; Cosgarea, M. Comparative analysis of GSTM1/GSTT1 null alleles and Ile105Val GSTP1 variant in patients with Nasal Polyposis and hyposmia in a Romanian population group. Rom. Rev. Lab. Med. 2013, 21, 189–196. [Google Scholar] [CrossRef]
- Dżaman, K.; Zagor, M.; Sarnowska, E.; Krzeski, A.; Kantor, I. The correlation of TAS2R38 gene variants with higher risk for chronic rhinosinusitis in Polish patients. Otolaryngol. Pol. 2016, 70, 13–18. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Moylan, B.; Leopold, D.A.; Kim, J.; Rubenstein, R.C.; Togias, A.; Proud, D.; Zeitlin, P.L.; Cutting, G.R. Mutation in the gene responsible for cystic fibrosis and predisposition to chronic rhinosinusitis in the general population. JAMA 2000, 284, 1814–1819. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Kim, J.; McWilliams, R.; Cutting, G.R. Increased prevalence of chronic rhinosinusitis in carriers of a cystic fibrosis mutation. Arch. Otolaryngol. Head Neck Surg. 2005, 131, 237–240. [Google Scholar] [CrossRef] [PubMed]
- Pinto, J.M.; Hayes, M.G.; Schneider, D.; Naclerio, R.M.; Ober, C. A genomewide screen for chronic rhinosinusitis genes identifies a locus on chromosome 7q. Laryngoscope 2008, 118, 2067–2072. [Google Scholar] [CrossRef]
- Yong, M.; Hernaiz-Leonardo, J.C.; Alqunaee, M.; Quon, B.S.; Javer, A. The prevalence of CFTR mutations in patients with chronic rhinosinusitis: A systematic review and meta-analysis. Clin. Otolaryngol. 2022, 47, 24–33. [Google Scholar] [CrossRef] [PubMed]
- Chang, E.H.; Tang, X.X.; Shah, V.S.; Launspach, J.L.; Ernst, S.E.; Hilkin, B.; Karp, P.H.; Alaiwa, M.H.A.; Graham, S.M.; Hornick, D.B.; et al. Medical reversal of chronic sinusitis in a cystic fibrosis patient with ivacaftor. Int. Forum Allergy Rhinol. 2016, 5, 178–181. [Google Scholar] [CrossRef] [PubMed]
- Al-Shemari, H.; Bossé, Y.; Hudson, T.J.; Cabaluna, M.; Duval, M.; Lemire, M.; Vallee-Smedja, S.; Frenkiel, S.; Desrosiers, M. Influence of leukotriene gene polymorphisms on chronic rhinosinusitis. BMC Med. Genet. 2008, 9, 21. [Google Scholar] [CrossRef] [PubMed]
- Pescador, D.B.; Isidoro-García, M.; García-Solaesa, V.; De Pedro, M.P.; Sanz, C.; Hernández-Hernández, L.; Sánchez-López, J.; Lorente, S.F.; Picado, C.; Valero, A.; et al. Genetic association study in nasal polyposis. J. Investig. Allergol. Clin. Immunol. 2012, 22, 331–340. [Google Scholar]
- Sitarek, P.; Zielinska-Blizniewska, H.; Dziki, L.; Milonski, J.; Przybylowska, K.; Mucha, B.; Olszewski, J.; Majsterek, I. Association of the -14C/G MET and the -765G/C COX-2 gene polymorphisms with the risk of chronic rhinosinusitis with nasal polyps in a Polish population. DNA Cell Biol. 2012, 31, 1258–1266. [Google Scholar] [CrossRef] [PubMed]
- Ober, C.; Yao, T. The genetics of asthma and allergic disease: A 21st century perspective. Immunol. Rev. 2011, 242, 10–30. [Google Scholar] [CrossRef] [PubMed]
- Salter, S.J.; Cox, M.J.; Turek, E.M.; Calus, S.T.; Cookson, W.O.; Moffatt, M.F.; Turner, P.; Parkhill, J.; Loman, N.J.; Walker, A.W. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014, 12, 87. [Google Scholar] [CrossRef] [PubMed]
- Tap, J.; Cools-Portier, S.; Pavan, S.; Druesne, A.; Öhman, L.; Törnblom, H.; Simren, M.; Derrien, M. Effects of the long-term storage of human fecal microbiota samples collected in RNAlater. Sci. Rep. 2019, 9, 601. [Google Scholar] [CrossRef]
- Turner, M.L.; Schnorfeil, F.M.; Brocker, T. MicroRNAs regulate dendritic cell differentiation and function. J. Immunol. 2011, 187, 3911–3917. [Google Scholar] [CrossRef]
- Ma, Z.; Shen, Y.; Zeng, Q.; Liu, J.; Yang, L.; Fu, R.; Hu, G. MiR-150-5p regulates EGR2 to promote the development of chronic rhinosinusitis via the DC-Th axis. Int. Immunopharmacol. 2018, 54, 188–197. [Google Scholar] [CrossRef]
- Agency USEP. Criteria Air Pollutants. 22 March 2021. Available online: https://www.epa.gov/criteria-air-pollutants (accessed on 9 February 2022).
- Leland, E.M.; Vohra, V.; Seal, S.M.; Zhang, Z.; Ramanathan, M. Environmental air pollution and chronic rhinosinusitis: A systematic review. Laryngoscope Investig. Otolaryngol. 2022, 7, 349–360. [Google Scholar] [CrossRef]
- London, N.R.; Tharakan, A.; Rule, A.M.; Lane, A.P.; Biswal, S.; Ramanathan, M. Air pollutant–mediated disruption of sinonasal epithelial cell barrier function is reversed by activation of the Nrf2 pathway. J. Allergy Clin. Immunol. 2016, 138, 1736–1738.e4. [Google Scholar] [CrossRef]
- De Schryver, E.; Derycke, L.; Campo, P.; Gabriels, E.; Joos, G.F.; Van Zele, T.; Bachert, C.; Hellings, P.W.; Gevaert, P. Alcohol hyper-responsiveness in chronic rhinosinusitis with nasal polyps. Clin. Exp. Allergy 2017, 47, 245–253. [Google Scholar] [CrossRef]
- Leru, P.M. Eosinophilic disorders: Evaluation of current classification and diagnostic criteria, proposal of a practical diagnostic algorithm. Clin. Transl. Allergy 2019, 9, 36. [Google Scholar] [CrossRef]
- Tong, J.; Gu, Q. Expression and Clinical Significance of Mucin Gene in Chronic Rhinosinusitis. Curr. Allergy Asthma Rep. 2020, 20, 63. [Google Scholar] [CrossRef]
- Tewfik, M.A.; Latterich, M.; DiFalco, M.R.; Samaha, M. Proteomics of nasal mucus in chronic rhinosinusitis. Am. J. Rhinol. 2007, 21, 680–685. [Google Scholar] [CrossRef]
- Badaai, Y.; DiFalco, M.; Tewfik, M.; Samaha, M. Quantitative Proteomics of Nasal Mucus in Chronic Sinusitis with Nasal Polyposis. J. Otolaryngol. Head Neck Surg. 2009, 38, 381–389. [Google Scholar] [PubMed]
- Fazlollahi, F.; Kongmanas, K.; Tanphaichitr, N.; Clair, J.M.; Gopen, Q.; Faull, K.F.; Suh, J.D. Lipidomic profiling of sinus mucosa from patients with chronic rhinosinusitis. Clin. Transl. Sci. 2015, 8, 107–115. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.S.; Kim, R.; Douglas, R. Is there a role for antibiotics in the treatment of chronic rhinosinusitis? J. Allergy Clin. Immunol. 2022, 149, 1504–1512. [Google Scholar] [CrossRef] [PubMed]
- Arcimowicz, M.; Niemczyk, K. EPOS 2020: What’s new for physician practitioners? Otolaryngol. Pol. 2020, 9, 7–17. [Google Scholar] [CrossRef]
- Barshak, M.B.; Durand, M.L. The Role of Infection and Antibiotics in Chronic Rhinosinusitis. Laryngoscope Investig. Otolaryngol. 2017, 2, 36–42. [Google Scholar] [CrossRef]
- Bose, S.; Grammer, L.C.; Peters, A.T. Infectious Chronic Rhinosinusitis. J. Allergy Clin. Immunol. Pract. 2016, 4, 584–589. [Google Scholar] [CrossRef]
- Michalik, M.; Podbielska-Kubera, A.; Samet, A.; Konopa, W. Multidrug-resistant strains of cogulase-negative staphylococci isolated from patients with chronic sinusitis—MDR, XDR, PDR strains. Otolaryngol. Pol. 2019, 74, 36–41. [Google Scholar] [CrossRef]
- Lux, C.A.; Mackenzie, B.W.; Johnston, J.; Zoing, M.; Biswas, K.; Taylor, M.W.; Douglas, R.G. Antibiotic Treatment for Chronic Rhinosinusitis: Prescription Patterns and Associations With Patient Outcome and the Sinus Microbiota. Front. Microbiol. 2020, 11, 595555. [Google Scholar] [CrossRef]
- Liu, C.M.; Soldanova, K.; Nordstrom, L.; Dwan, M.G.; Moss, O.L.; Contente-Cuomo, T.L.; Keim, P.; Price, L.B.; Lane, A.P. Medical therapy reduces microbiota diversity and evenness in surgically recalcitrant chronic rhinosinusitis. Int. Forum Allergy Rhinol. 2013, 3, 775–781. [Google Scholar] [CrossRef]
- Feazel, L.M.; Frank, D.N.; Ramakrishnan, V.R. Update on bacterial detection methods in chronic rhinosinusitis: Implications for clinicians and research scientists. Int. Forum Allergy Rhinol. 2011, 1, 451–459. [Google Scholar] [CrossRef] [PubMed]
- Abreu, N.A.; Nagalingam, N.A.; Song, Y.; Roediger, F.C.; Pletcher, S.D.; Goldberg, A.N.; Lynch, S.V. Sinus microbiome diversity depletion and Corynebacterium tuberculostearicum enrichment mediates rhinosinusitis. Sci. Transl. Med. 2012, 4, 151ra124. [Google Scholar] [CrossRef] [PubMed]
- Uehara, Y.; Nakama, H.; Agematsu, K.; Uchida, M.; Kawakami, Y.; Fattah, A.A.; Maruchi, N. Bacterial interference among nasal inhabitants: Eradication of Staphylococcus aureus from nasal cavities by artificial implantation of Corynebacterium sp. J. Hosp. Infect. 2000, 44, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Jurkiewicz, D.; Kupczyk, M.; Brożek-Mądry, E.; Rapiejko, P. Biologicals in the treatment of chronic rhinosinusitis with nasal polyps—Position of the Polish Society of Otorhinolaryngologists—Head and Neck Surgeons and the Polish Society of Allergology expert. Otolaryngol. Pol. 2023, 77, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Czerwaty, K.; Miechowski, W.; Godlewska, I.; Dżaman, K. Biological treatment in chronic rhinosinusitis: The current state of knowledge. Otolaryngol. Pol. 2022, 11, 22–28. [Google Scholar] [CrossRef]
- Tai, J.; Han, M.; Kim, T.H. Therapeutic Strategies of Biologics in Chronic Rhinosinusitis: Current Options and Future Targets. Int. J. Mol. Sci. 2022, 23, 5523. [Google Scholar] [CrossRef]
- Laidlaw, T.M.; Bachert, C.; Amin, N.; Desrosiers, M.; Hellings, P.W.; Mullol, J.; Maspero, J.F.; Gevaert, P.; Zhang, M.; Mao, X.; et al. Dupilumab improves upper and lower airway disease control in chronic rhinosinusitis with nasal polyps and asthma. Ann. Allergy Asthma Immunol. 2021, 126, 584–592.e1. [Google Scholar] [CrossRef]
- Wijs, L.; Bosma, A.; Erler, N.; Hollestein, L.; Gerbens, L.; Middelkamp-Hup, M.; Kunkeler, A.; Nijsten, T.; Spuls, P.; Hijnen, D. Effectiveness of dupilumab treatment in 95 patients with atopic dermatitis: Daily practice data. Br. J. Dermatol. 2020, 182, 418–426. [Google Scholar] [CrossRef]
- Iribarren, C.; Rahmaoui, A.; Long, A.A.; Szefler, S.J.; Bradley, M.S.; Carrigan, G.; Eisner, M.D.; Chen, H.; Omachi, T.A.; Farkouh, M.E.; et al. Cardiovascular and cerebrovascular events among patients receiving omalizumab: Results from EXCELS, a prospective cohort study in moderate to severe asthma. J. Allergy Clin. Immunol. 2017, 139, 1489–1495.e5. [Google Scholar] [CrossRef]
- Gevaert, P.; Omachi, T.A.; Corren, J.; Mullol, J.; Han, J.; Lee, S.E.; Kaufman, D.; Ligueros-Saylan, M.; Howard, M.; Zhu, R.; et al. Efficacy and safety of omalizumab in nasal polyposis: 2 randomized phase 3 trials. J. Allergy Clin. Immunol. 2020, 146, 595–605. [Google Scholar] [CrossRef] [PubMed]
- Gevaert, P.; Calus, L.; Van Zele, T.; Blomme, K.; De Ruyck, N.; Bauters, W.; Hellings, P.; Brusselle, G.; De Bacquer, D.; van Cauwenberge, P.; et al. Omalizumab is effective in allergic and nonallergic patients with nasal polyps and asthma. J. Allergy Clin. Immunol. 2013, 131, 110–116.e1. [Google Scholar] [CrossRef]
- Bidder, T.; Sahota, J.; Rennie, C.; Lund, V.J.; Robinson, D.S.; Kariyawasam, H.H. Omalizumab treats chronic rhinosinusitis with nasal polyps and asthma together—A real life study. Rhinology 2018, 56, 42–45. [Google Scholar] [CrossRef] [PubMed]
- Armengot-Carceller, M.; Gómez-Gómez, M.J.; García-Navalón, C.; Doménech-Campos, E.; Muñoz-Fernández, N.; de Miguel, A.G.-L.; Marco-Algarra, J.; Palop-Cervera, M.; Piñero, A.G. Effects of Omalizumab Treatment in Patients With Recalcitrant Nasal Polyposis and Mild Asthma: A Multicenter Retrospective Study. Am. J. Rhinol. Allergy 2021, 35, 516–524. [Google Scholar] [CrossRef]
- Broughton, S.E.; Nero, T.L.; Dhagat, U.; Kan, W.L.; Hercus, T.R.; Tvorogov, D.; Lopez, A.F.; Parker, M.W. The βc receptor family—Structural insights and their functional implications. Cytokine 2015, 74, 247–258. [Google Scholar] [CrossRef] [PubMed]
- Gevaert, P.; Van Bruaene, N.; Cattaert, T.; Van Steen, K.; Van Zele, T.; Acke, F.; De Ruyck, N.; Blomme, K.; Sousa, A.R.; Marshall, R.P.; et al. Mepolizumab, a humanized anti–IL-5 mAb, as a treatment option for severe nasal polyposis. J. Allergy Clin. Immunol. 2011, 128, 989–995.e8. [Google Scholar] [CrossRef]
- Bachert, C.; Sousa, A.R.; Lund, V.J.; Scadding, G.K.; Gevaert, P.; Nasser, S.; Durham, S.R.; Cornet, M.E.; Kariyawasam, H.H.; Gilbert, J.; et al. Reduced need for surgery in severe nasal polyposis with mepolizumab: Randomized trial. J. Allergy Clin. Immunol. 2017, 140, 1024–1031.e14. [Google Scholar] [CrossRef] [PubMed]
- Whittington, M.D.; McQueen, R.B.; Ollendorf, D.A.; Tice, J.A.; Chapman, R.H.; Pearson, S.D.; Campbell, J.D. Assessing the value of mepolizumab for severe eosinophilic asthma: A cost-effectiveness analysis. Ann. Allergy Asthma Immunol. 2017, 118, 220–225. [Google Scholar] [CrossRef] [PubMed]
- Gevaert, P.; Langloidolt, D.; Lackner, A.; Stammberger, H.; Staudinger, H.; Vanzele, T.; Holtappels, G.; Tavernier, J.; Vancauwenberge, P.; Bachert, C. Nasal IL-5 levels determine the response to anti–IL-5 treatment in patients with nasal polyps. J. Allergy Clin. Immunol. 2006, 118, 1133–1141. [Google Scholar] [CrossRef] [PubMed]
- De Corso, E.; Bellocchi, G.; De Benedetto, M.; Lombardo, N.; Macchi, A.; Malvezzi, L.; Motta, G.; Pagella, F.; Vicini, C.; Passali, D. Biologics for severe uncontrolled chronic rhinosinusitis with nasal polyps: A change management approach. Consensus of the Joint Committee of Italian Society of Otorhinolaryngology on biologics in rhinology. Acta Otorhinolaryngol. Ital. 2022, 42, 1–16. [Google Scholar] [CrossRef]
- Harrison, T.W.; Chanez, P.; Menzella, F.; Canonica, G.W.; Louis, R.; Cosio, B.G.; Lugogo, N.L.; Mohan, A.; Burden, A.; McDermott, L.; et al. Onset of effect and impact on health-related quality of life, exacerbation rate, lung function, and nasal polyposis symptoms for patients with severe eosinophilic asthma treated with benralizumab (ANDHI): A randomised, controlled, phase 3b trial. Lancet Respir. Med. 2021, 9, 260–274. [Google Scholar] [CrossRef]
- Haloob, N.; Karamali, K.; Hopkins, C. The Role of Biologics in the Treatment of Chronic Rhinosinusitis. BioDrugs 2023, 37, 477–487. [Google Scholar] [CrossRef]



| Techniques/Methods | Advantages | Disadvantages |
|---|---|---|
| Culture techniques |
|
|
| Molecular methods |
|
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Michalik, M.; Krawczyk, B. Chronic Rhinosinusitis—Microbiological Etiology, Potential Genetic Markers, and Diagnosis. Int. J. Mol. Sci. 2024, 25, 3201. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms25063201
Michalik M, Krawczyk B. Chronic Rhinosinusitis—Microbiological Etiology, Potential Genetic Markers, and Diagnosis. International Journal of Molecular Sciences. 2024; 25(6):3201. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms25063201
Chicago/Turabian StyleMichalik, Michał, and Beata Krawczyk. 2024. "Chronic Rhinosinusitis—Microbiological Etiology, Potential Genetic Markers, and Diagnosis" International Journal of Molecular Sciences 25, no. 6: 3201. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms25063201