The Evolution and Biogeography of Wolbachia in Ants (Hymenoptera: Formicidae)
Abstract
:1. Introduction
2. Material and Methods
2.1. Phylogenetic Reconstruction
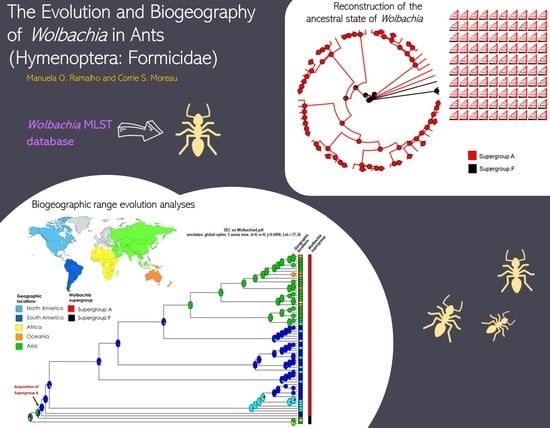
2.2. Reconstruction of the Ancestral State of Wolbachia
2.3. Biogeographic Range Evolution Analyses
3. Results
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kikuchi, Y.; Hosokawa, T.; Fukatsu, T. Insect-microbe mutualism without vertical transmission: A stinkbug acquires a beneficial gut symbiont from the environment every generation. Appl. Environ. Microbiol. 2007, 73, 4308–4316. [Google Scholar] [CrossRef] [Green Version]
- Buchner, P. Endosymbiosis of Animals with Plant Microorganisms.1965.-Google Acadêmico; Buchner, P., Ed.; Interscience Publishers: New York, NY, USA, 1965. [Google Scholar]
- Bourtzis, K.; Miller, T. Insect Symbiosis; CRC Press: Boca Ranton, FL, USA, 2006; Volume 2. [Google Scholar]
- Hu, Y.; Sanders, J.G.; Łukasik, P.; D’Amelio, C.L.; Millar, J.S.; Vann, D.R.; Lan, Y.; Newton, J.A.; Schotanus, M.; Kronauer, D.J.C.; et al. Herbivorous turtle ants obtain essential nutrients from a conserved nitrogen-recycling gut microbiome. Nat. Commun. 2018, 9, 964. [Google Scholar] [CrossRef] [Green Version]
- Saraithong, P.; Li, Y.; Saenphet, K.; Chen, Z.; Chantawannakul, P. Midgut bacterial communities in the giant Asian honeybee (Apis dorsata) across 4 developmental stages: A comparative study. Insect Sci. 2017, 24, 81–92. [Google Scholar] [CrossRef]
- Russell, J.A.; Funaro, C.F.; Giraldo, Y.M.; Goldman-Huertas, B.; Suh, D.; Kronauer, D.J.C.; Moreau, C.S.; Pierce, N.E. A Veritable Menagerie of Heritable Bacteria from Ants, Butterflies, and Beyond: Broad Molecular Surveys and a Systematic Review. PLoS ONE 2012, 7, e51027. [Google Scholar] [CrossRef]
- Hilgenboecker, K.; Hammerstein, P.; Schlattmann, P.; Telschow, A.; Werren, J.H. How many species are infected with Wolbachia? A statistical analysis of current data. FEMS Microbiol. Lett. 2008, 281, 215–220. [Google Scholar] [CrossRef] [Green Version]
- Werren, J.H.; Baldo, L.; Clark, M.E. Wolbachia: Master manipulators of invertebrate biology. Nat. Rev. Microbiol. 2008, 6, 741–751. [Google Scholar] [CrossRef]
- Russell, J.A.; Goldman-Huertas, B.; Moreau, C.S.; Baldo, L.; Stahlhut, J.K.; Werren, J.H.; Pierce, N.E. Specialization and geographic isolation among Wolbachia symbionts from ants and lycaenid butterflies. Evolution 2009, 63, 624–640. [Google Scholar] [CrossRef]
- Lefoulon, E.; Clark, T.; Borveto, F.; Perriat-Sanguinet, M.; Moulia, C.; Slatko, B.E.; Gavotte, L. Pseudoscorpion Wolbachia symbionts: Diversity and evidence for a new supergroup S. BMC Microbiol. 2020, 20, 188. [Google Scholar] [CrossRef]
- Baldo, L.; Werren, J.H. Revisiting Wolbachia supergroup typing based on WSP: Spurious lineages and discordance with MLST. Curr. Microbiol. 2007, 55, 81–87. [Google Scholar] [CrossRef]
- Gerth, M. Classification of Wolbachia (Alphaproteobacteria, Rickettsiales): No evidence for a distinct supergroup in cave spiders. Infect. Genet. Evol. 2016, 43, 378–380. [Google Scholar] [CrossRef]
- Bordenstein, S.; Rosengaus, R.B. Discovery of a novel Wolbachia supergroup in isoptera. Curr. Microbiol. 2005, 51, 393–398. [Google Scholar] [CrossRef] [PubMed]
- Glowska, E.; Dragun-Damian, A.; Dabert, M.; Gerth, M. New Wolbachia supergroups detected in quill mites (Acari: Syringophilidae). Infect. Genet. Evol. 2015, 30, 140–146. [Google Scholar] [CrossRef] [PubMed]
- Lo, N.; Casiraghi, M.; Salati, E.; Bazzocchi, C.; Bandi, C. How Many Wolbachia Supergroups Exist? Mol. Biol. Evol. 2002, 19, 341–346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lo, N.; Paraskevopoulos, C.; Bourtzis, K.; O’Neill, S.L.; Werren, J.H.; Bordenstein, S.R.; Bandi, C. Taxonomic status of the intracellular bacterium Wolbachia pipientis. Int. J. Syst. Evol. Microbiol. 2007, 57, 654–657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ros, V.I.D.; Fleming, V.M.; Feil, E.J.; Breeuwer, J.A.J. How diverse is the genus Wolbachia? Multiple-gene sequencing reveals a putatively new Wolbachia supergroup recovered from spider mites (Acari: Tetranychidae). Appl. Environ. Microbiol. 2009, 75, 1036–1043. [Google Scholar] [CrossRef] [Green Version]
- Ferri, E.; Bain, O.; Barbuto, M.; Martin, C.; Lo, N.; Uni, S.; Landmann, F.; Baccei, S.G.; Guerrero, R.; de Souza Lima, S.; et al. New Insights into the Evolution of Wolbachia Infections in Filarial Nematodes Inferred from a Large Range of Screened Species. PLoS ONE 2011, 6, e20843. [Google Scholar] [CrossRef] [Green Version]
- Lefoulon, E.; Gavotte, L.; Junker, K.; Barbuto, M.; Uni, S.; Landmann, F.; Laaksonen, S.; Saari, S.; Nikander, S.; de Souza Lima, S.; et al. A new type F Wolbachia from Splendidofilariinae (Onchocercidae) supports the recent emergence of this supergroup. Int. J. Parasitol. 2012, 42, 1025–1036. [Google Scholar] [CrossRef]
- Bolton, B. An Online Catalog of the Ants of the World. Available online: http://www.antcat.org/ (accessed on 20 October 2016).
- Singh, R.; Linksvayer, T.A. Wolbachia-infected ant colonies have increased reproductive investment and an accelerated life cycle. J. Exp. Biol. 2020, 223. [Google Scholar] [CrossRef]
- Cheng, D.; Chen, S.; Huang, Y.; Pierce, N.E.; Riegler, M.; Yang, F.; Zeng, L.; Lu, Y.; Liang, G.; Xu, Y. Symbiotic microbiota may reflect host adaptation by resident to invasive ant species. PLoS Pathog. 2019, 15, e1007942. [Google Scholar] [CrossRef] [Green Version]
- Fernando de Souza, R.; Daivison Silva Ramalho, J.; Santina de Castro Morini, M.; Wolff, J.L.C.; Araújo, R.C.; Mascara, D. Identification and Characterization of Wolbachia in Solenopsis saevissima Fire Ants (Hymenoptera: Formicidae) in Southeastern Brazil. Curr. Microbiol. 2009, 58, 189–194. [Google Scholar] [CrossRef]
- Frost, C.L.; Fernandez-Marin, H.; Smith, J.E.; Hughes, W.O.H. Multiple gains and losses of Wolbachia symbionts across a tribe of fungus-growing ants. Mol. Ecol. 2010, 19, 4077–4085. [Google Scholar] [CrossRef]
- Frost, C.L.; Pollock, S.W.; Smith, J.E.; Hughes, W.O.H. Wolbachia in the flesh: Symbiont intensities in germ-line and somatic tissues challenge the conventional view of Wolbachia transmission routes. PLoS ONE 2014, 9, e95122. [Google Scholar] [CrossRef] [Green Version]
- Martins, C.; Souza, R.F.; Bueno, O.C. Presence and distribution of the endosymbiont Wolbachia among Solenopsis spp. (Hymenoptera: Formicidae) from Brazil and its evolutionary history. J. Invertebr. Pathol. 2012, 109, 287–296. [Google Scholar] [CrossRef] [Green Version]
- Kautz, S.; Rubin, B.E.R.; Moreau, C.S. Bacterial Infections across the Ants: Frequency and Prevalence of Wolbachia, Spiroplasma, and Asaia. Psyche A J. Entomol. 2013, 2013, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Ramalho, M.O.; Martins, C.; Silva, L.M.R.; Martins, V.G.; Bueno, O.C. Intracellular symbiotic bacteria of Camponotus textor, Forel (Hymenoptera, Formicidae). Curr. Microbiol. 2017, 74, 589–597. [Google Scholar] [CrossRef] [Green Version]
- Ramalho, M.O.; Vieira, A.S.; Pereira, M.C.; Moreau, C.S.; Bueno, O.C. Transovarian Transmission of Blochmannia and Wolbachia Endosymbionts in the Neotropical Weaver Ant Camponotus textor (Hymenoptera, Formicidae). Curr. Microbiol. 2018, 75, 866–873. [Google Scholar] [CrossRef]
- Ramalho, M.O.; Bueno, O.C.; Moreau, C.S. Species-specific signatures of the microbiome from Camponotus and Colobopsis ants across developmental stages. PLoS ONE 2017, 12, e0187461. [Google Scholar] [CrossRef] [Green Version]
- Ramalho, M.O.; Bueno, O.C.; Moreau, C.S. Microbial composition of spiny ants (Hymenoptera: Formicidae: Polyrhachis) across their geographic range. BMC Evol. Biol. 2017, 17, 96. [Google Scholar] [CrossRef] [Green Version]
- Tseng, S.P.; Wetterer, J.K.; Suarez, A.V.; Lee, C.Y.; Yoshimura, T.; Shoemaker, D.W.; Yang, C.C.S. Genetic Diversity and Wolbachia Infection Patterns in a Globally Distributed Invasive Ant. Front. Genet. 2019, 10, 838. [Google Scholar] [CrossRef]
- Kelly, M.; Price, S.L.; de Oliveira Ramalho, M.; Moreau, C.S. Diversity of Wolbachia Associated with the Giant Turtle Ant, Cephalotes atratus. Curr. Microbiol. 2019, 76, 1330–1337. [Google Scholar] [CrossRef]
- Ramalho, M.O.; Moreau, C.S.; Bueno, O.C. The Potential Role of Environment in Structuring the Microbiota of Camponotus across Parts of the Body. Adv. Entomol. 2019, 7, 47–70. [Google Scholar] [CrossRef] [Green Version]
- Reeves, D.D.; Price, S.L.; Ramalho, M.O.; Moreau, C.S. The Diversity and Distribution of Wolbachia, Rhizobiales, and Ophiocordyceps Within the Widespread Neotropical Turtle Ant, Cephalotes atratus (Hymenoptera: Formicidae). Neotrop. Entomol. 2020, 49, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Baldo, L.; Lo, N.; Werren, J.H. Mosaic nature of the wolbachia surface protein. J. Bacteriol. 2005, 187, 5406–5418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paraskevopoulos, C.; Bordenstein, S.; Wernegreen, J.; Werren, J.; Bourtzis, K. Toward a Wolbachia multilocus sequence typing system: Discrimination of Wolbachia strains present in Drosophila species. Curr. Microbiol. 2006, 53, 388–395. [Google Scholar] [CrossRef]
- Baldo, L.; Dunning Hotopp, J.C.; Jolley, K.A.; Bordenstein, S.R.; Biber, S.A.; Choudhury, R.R.; Hayashi, C.; Maiden, M.C.J.; Tettelin, H.; Werren, J.H. Multilocus sequence typing system for the endosymbiont Wolbachia pipientis. Appl. Environ. Microbiol. 2006, 72, 7098–7110. [Google Scholar] [CrossRef] [Green Version]
- Ali, H.; Muhammad, A.; Hou, Y. Infection Density Dynamics and Phylogeny of Wolbachia Associated with Coconut Hispine Beetle, Brontispa longissima (Gestro) (Coleoptera: Chrysomelidae), by Multilocus Sequence Type. Artic. J. Microbiol. Biotechnol. 2018, 28, 796–808. [Google Scholar] [CrossRef] [Green Version]
- Ali, H.; Muhammad, A.; Sanda Bala, N.; Hou, Y. The Endosymbiotic Wolbachia and Host COI Gene Enables to Distinguish Between Two Invasive Palm Pests; Coconut Leaf Beetle, Brontispa longissima and Hispid Leaf Beetle, Octodonta nipae. J. Econ. Entomol. 2018, 111, 2894–2902. [Google Scholar] [CrossRef] [Green Version]
- Betelman, K.; Caspi-Fluger, A.; Shamir, M.; Chiel, E. Identification and characterization of bacterial symbionts in three species of filth fly parasitoids. FEMS Microbiol. Ecol. 2017, 93. [Google Scholar] [CrossRef]
- Prezotto, L.F.; Perondini, A.L.P.; Hernández-Ortiz, V.; Marino, C.L.; Selivon, D. Wolbachia strains in cryptic species of the Anastrepha fraterculus complex (Diptera, Tephritidae) along the Neotropical Region. Syst. Appl. Microbiol. 2017, 40, 59–67. [Google Scholar] [CrossRef] [Green Version]
- Higgins, D.; Bleasby, A.; Fuchs, R. CLUSTAL V: Improved software for multiple sequence alignment. Comput. Appl. 1992, 8, 189–191. [Google Scholar]
- Hall, T. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New Methods for Selecting Partitioned Models of Evolution for Molecular and Morphological Phylogenetic Analyses. Mol. Biol. Evol. 2016, 34, msw260. [Google Scholar] [CrossRef] [Green Version]
- Huelsenbeck, J.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the 2010 Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; pp. 1–8. [Google Scholar]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. The CIPRES Portals. Available online: http://www.phylo.org/ (accessed on 20 September 2020).
- Paradis, E.; Claude, J.; Strimmer, K. APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics 2004, 20, 289–290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020; Available online: http://www.R-project.org/ (accessed on 20 September 2020).
- Kim, J.; Sanderson, M.J. Penalized Likelihood Phylogenetic Inference: Bridging the Parsimony-Likelihood Gap. Syst. Biol. 2008, 57, 665–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paradis, E. Molecular dating of phylogenies by likelihood methods: A comparison of models and a new information criterion. Mol. Phylogenet. Evol. 2013, 67, 436–444. [Google Scholar] [CrossRef] [Green Version]
- Sanderson, M.J. Estimating Absolute Rates of Molecular Evolution and Divergence Times: A Penalized Likelihood Approach. Mol. Biol. Evol. 2002, 19, 101–109. [Google Scholar] [CrossRef] [Green Version]
- Pagel, M. Detecting correlated evolution on phylogenies: A general method for the comparative analysis of discrete characters. Proc. R. Soc. London. Ser. B Biol. Sci. 1994, 255, 37–45. [Google Scholar] [CrossRef]
- Schluter, D.; Price, T.; Mooers, A.Ø.; Ludwig, D. Likelihood of ancestor states in adaptive radiation. Evolution 1997, 51, 1699–1711. [Google Scholar] [CrossRef]
- Maintainer, L.J.R.; Revell, L.J. Package “Phytools” Title Phylogenetic Tools for Comparative Biology (and Other Things). R Package. 2020. Available online: https://0-besjournals-onlinelibrary-wiley-com.brum.beds.ac.uk/doi/full/10.1111/j.2041-210X.2011.00169.x (accessed on 12 November 2020).
- Huelsenbeck, J.P.; Nielsen, R.; Bollback, J.P. Stochastic Mapping of Morphological Characters. Syst. Biol. 2003, 52, 131–158. [Google Scholar] [CrossRef]
- Bollback, J.P. SIMMAP: Stochastic character mapping of discrete traits on phylogenies. BMC Bioinf. 2006, 7, 88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matzke, N.J. BioGeoBEARS: An R package for model testing and ancestral state estimation in historical biogeography. Methods Ecol. Evol. 2014. [Google Scholar]
- Matzke, N.J. Model selection in historical biogeography reveals that founder-event speciation is a crucial process in island clades. Syst. Biol. 2014, 63, 951–970. [Google Scholar] [CrossRef] [PubMed]
- Ree, R.H.; Smith, S.A. Maximum Likelihood Inference of Geographic Range Evolution by Dispersal, Local Extinction, and Cladogenesis. Syst. Biol. 2008, 57, 4–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fuentes-Castillo, T.; Hernández, H.J.; Pliscoff, P. Hotspots and ecoregion vulnerability driven by climate change velocity in Southern South America. Reg. Environ. Chang. 2020, 20, 1–15. [Google Scholar] [CrossRef]
- Cantidio, L.S.; Souza, A.F. Aridity, soil and biome stability influence plant ecoregions in the Atlantic Forest, a biodiversity hotspot in South America. Ecography 2019, 42, 1887–1898. [Google Scholar] [CrossRef] [Green Version]
- Martins, C.; Correa Bueno, O. Determinação de Cepas de Wolbachia em Populações Naturais de Solenopsis spp. (Hymenoptera: Formicidae) Analisadas via Multilocus Sequence Typing (MLST): Diversidade Genética, Coevolução e Recombinação; Universidade Estadual Paulista (UNESP): Rio Claro, Brazil, 2014. [Google Scholar]
- Comandatore, F.; Sassera, D.; Montagna, M.; Kumar, S.; Koutsovoulos, G.; Thomas, G.; Repton, C.; Babayan, S.A.; Gray, N.; Cordaux, R.; et al. Phylogenomics and Analysis of Shared Genes Suggest a Single Transition to Mutualism in Wolbachia of Nematodes. Genome Biol. Evol. 2013, 5, 1668–1674. [Google Scholar] [CrossRef] [Green Version]
- Narita, S.; Shimajiri, Y.; Nomura, M. Strong cytoplasmic incompatibility and high vertical transmission rate can explain the high frequencies of Wolbachia infection in Japanese populations of Colias erate poliographus (Lepidoptera: Pieridae). Bull. Entomol. Res. 2009, 99, 385–391. [Google Scholar] [CrossRef]
- Gerth, M.; Röthe, J.; Bleidorn, C. Tracing horizontal Wolbachia movements among bees (Anthophila): A combined approach using multilocus sequence typing data and host phylogeny. Mol. Ecol. 2013, 22, 6149–6162. [Google Scholar] [CrossRef]
- Duplouy, A.; Couchoux, C.; Hanski, I.; van Nouhuys, S. Wolbachia Infection in a Natural Parasitoid Wasp Population. PLoS ONE 2015, 10, e0134843. [Google Scholar] [CrossRef] [Green Version]
- Bouwma, A.M.; Shoemaker, D. Wolbachia wSinvictaA Infections in Natural Populations of the Fire Ant Solenopsis invicta: Testing for Phenotypic Effects. J. Insect Sci. 2011, 11, 1–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tolley, S.J.A.; Nonacs, P.; Sapountzis, P. Wolbachia Horizontal Transmission Events in Ants: What Do We Know and What Can We Learn? Front. Microbiol. 2019, 10, 296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmed, M.Z.; Araujo-Jnr, E.V.; Welch, J.J.; Kawahara, A.Y. Wolbachia in butterflies and moths: Geographic structure in infection frequency. Front. Zool. 2015, 12, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmed, M.Z.; Breinholt, J.W.; Kawahara, A.Y. Evidence for common horizontal transmission of Wolbachia among butterflies and moths. BMC Evol. Biol. 2016, 16, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| id | Supergroup | Host Genus | Host Species | Locality | Submitter | Sequence Type (ST) |
|---|---|---|---|---|---|---|
| 2 | A | Solenopsis | invicta | Argentina | Laura Baldo | 29 |
| 4 | A | Camponotus | pennsylvanicus | USA | Laura Baldo | 33 |
| 103 | A | Formica | occulta | USA | Jacob Russell | 43 |
| 104 | A | Pseudomyrmex | apache | USA | Jacob Russell | 44 |
| 105 | A | Stenamma | snellingi | USA | Jacob Russell | 45 |
| 106 | A | Azteca | spp. | Ecuador | Jacob Russell | 46 |
| 107 | A | Wasmannia | spp. | Peru | Jacob Russell | 47 |
| 108 | A | Metapone | madagascaria | Madagascar | Jacob Russell | 48 |
| 109 | A | Myrmica | incompleta | USA | Jacob Russell | 49 |
| 110 | A | Polyergus | breviceps | USA | Jacob Russell | 50 |
| 111 | A | Technomyrmex | albipes | Philippines | Jacob Russell | 19 |
| 112 | A | Polyrhachis | vindex | Philippines | Jacob Russell | 51 |
| 113 | A | Anoplolepis | gracillipes | Philippines | Jacob Russell | 52 |
| 114 | A | Notoncus | spp. | Australia | Jacob Russell | 53 |
| 115 | A | Leptomyrmex | spp. | Australia | Jacob Russell | 19 |
| 116 | A | Myrmecorhynchus | spp. | Australia | Jacob Russell | 54 |
| 117 | A | Pheidole | minutula | French Guiana | Jacob Russell | 55 |
| 119 | A | Lophomyrmex | spp. | Thailand | Jacob Russell | 52 |
| 120 | A | Camponotus | leonardi | Thailand | Jacob Russell | 57 |
| 121 | A | Pheidole | vallicola | USA | Jacob Russell | 58 |
| 122 | A | Rhytidoponera | metallica | Australia | Jacob Russell | 59 |
| 124 | A | Pheidole | plagiara | Thailand | Jacob Russell | 19 |
| 125 | A | Pheidole | sauberi | Thailand | Jacob Russell | 19 |
| 126 | A | Pheidole | gatesi | Vietnam | Jacob Russell | 60 |
| 127 | A | Pheidole | spp. | Thailand | Jacob Russell | 61 |
| 129 | A | Dorymyrmex | elegans | USA | Jacob Russell | 63 |
| 134 | A | Odontomachus | clarus | USA | Jacob Russell | 111 |
| 135 | A | Ochetellus | glaber | Australia | Jacob Russell | 112 |
| 137 | A | Pheidole | coloradensis | USA | Jacob Russell | 114 |
| 138 | A | Pheidole | micula | USA | Jacob Russell | 115 |
| 139 | A | Pheidole | vistana | Mexico | Jacob Russell | 116 |
| 140 | A | Pheidole | obtusospinosa | USA | Jacob Russell | 117 |
| 141 | A | Pheidole | spp. | Indonesia | Jacob Russell | 118 |
| 143 | A | Aenictus | spp. | Thailand | Jacob Russell | 120 |
| 144 | A | Crematogaster | spp. | Thailand | Jacob Russell | 121 |
| 145 | A | Solenopsis | spp. | Thailand | Jacob Russell | 122 |
| 146 | A | Leptogenys | spp. | Thailand | Jacob Russell | 19 |
| 147 | A | Pheidole | planifrons | Thailand | Jacob Russell | 19 |
| 148 | A | Monomorium | chinense | Thailand | Jacob Russell | 123 |
| 149 | F | Ocymyrmex | picardi | Congo (DRC] | Jacob Russell | 124 |
| 558 | A | Camponotus | textor | Brazil | Manuela Ramalho | 347 |
| 1827 | A | Paratrechina | longicornis | Taiwan | Tseng ShuPing | 19 |
| 1828 | F | Paratrechina | longicornis | Taiwan | Tseng ShuPing | 471 |
| 1868 | A | Cephalotes | atratus | Brazil | Madeleine Kelly | 494 |
| 1869 | A | Cephalotes | atratus | French Guiana | Madeleine Kelly | 495 |
| 1870 | A | Cephalotes | atratus | Brazil | Madeleine Kelly | 496 |
| 1871 | A | Cephalotes | atratus | Guyana | Madeleine Kelly | 497 |
| 1872 | A | Cephalotes | atratus | Brazil | Madeleine Kelly | 498 |
| 1873 | A | Cephalotes | atratus | Peru | Madeleine Kelly | 499 |
| 1989 | A | Cardiocondyla | spp. | India | Manisha Gupta | 550 |
| 1996 | F | Paratrechina | spp. | India | Manisha Gupta | 557 |
| 2010 | A | Pheidole | spp. | India | Manisha Gupta | 571 |
| 2017 | A | Anoplolepis | gracillipes | Malaysia | Tseng ShuPing | 52 |
| 2022 | A | Camponotus | spp. | Malaysia | Tseng ShuPing | 576 |
| 2023 | A | Camponotus | spp. | Malaysia | Tseng ShuPing | 577 |
| Not defined | A | Solenopsis | spp. | Brazil | Cintia Martins | 314 |
| Not defined | A | Solenopsis | spp. | Brazil | Cintia Martins | 315 |
| Not defined | A | Solenopsis | spp. | Brazil | Cintia Martins | 316 |
| Not defined | A | Solenopsis | spp. | Brazil | Cintia Martins | 317 |
| Not defined | A | Solenopsis | spp. | Brazil | Cintia Martins | 318 |
| Not defined | A | Solenopsis | spp. | Brazil | Cintia Martins | 319 |
| Not defined | A | Solenopsis | spp. | Brazil | Cintia Martins | 320 |
| Not defined | A | Solenopsis | spp. | Brazil | Cintia Martins | 321 |
| Not defined | A | Solenopsis | spp. | Brazil | Cintia Martins | 322 |
| Not defined | A | Solenopsis | spp. | Brazil | Cintia Martins | 323 |
| Not defined | A | Solenopsis | spp. | Brazil | Cintia Martins | 324 |
| Not defined | A | Solenopsis | spp. | Brazil | Cintia Martins | 325 |
| Not defined | A | Solenopsis | spp. | Brazil | Cintia Martins | 326 |
| Not defined | A | Solenopsis | spp. | Brazil | Cintia Martins | 327 |
| Not defined | A | Solenopsis | spp. | Brazil | Cintia Martins | 328 |
| 59 | F | Opistophthalmus | chaperi | South Africa | Laura Baldo | 78 |
| Model | Likelihood Ratio Test (LRT) | Akaike Information Criterion (AIC) |
|---|---|---|
| DEC + J Model | −77.355 | 1.28 × 1013 |
| DEC Model | −108.541 | 7.77× 10−14 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramalho, M.O.; Moreau, C.S. The Evolution and Biogeography of Wolbachia in Ants (Hymenoptera: Formicidae). Diversity 2020, 12, 426. https://0-doi-org.brum.beds.ac.uk/10.3390/d12110426
Ramalho MO, Moreau CS. The Evolution and Biogeography of Wolbachia in Ants (Hymenoptera: Formicidae). Diversity. 2020; 12(11):426. https://0-doi-org.brum.beds.ac.uk/10.3390/d12110426
Chicago/Turabian StyleRamalho, Manuela O., and Corrie S. Moreau. 2020. "The Evolution and Biogeography of Wolbachia in Ants (Hymenoptera: Formicidae)" Diversity 12, no. 11: 426. https://0-doi-org.brum.beds.ac.uk/10.3390/d12110426