Phenotypic and Genotypic Comparison of Antimicrobial-Resistant Variants of Escherichia coli and Salmonella Typhimurium Isolated from Evolution Assays with Antibiotics or Commercial Products Based on Essential Oils
Abstract
:1. Introduction
2. Results and Discussion
2.1. Chemical Composition of AEN and COLIFIT
2.2. Minimum Inhibitory Concentration (MIC)
2.3. Mutagenesis Frequency
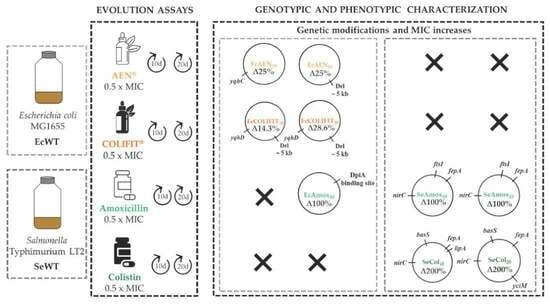
2.4. Phenotypic Characterization of Evolved Mutants
2.5. Genotypic Characterization of RVs
3. Materials and Methods
3.1. Antimicrobial Compounds and Chemical Analysis
3.2. Microorganisms and Growth Conditions
3.3. Minimum Inhibitory Concentration (MIC) and Minimum Bactericidal Concentration (MCB)
3.4. Mutagenesis Frequency
3.5. Evolution Assays
3.6. Phenotypic Characterization of Evolved Strains
3.7. Genotypic Characterization
3.8. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- European Medicines Agency. Sales of Veterinary Antimicrobial Agents in 31 European Countries in 2021. Trends from 2010 to 2021. Twelfth ESVAC Report. 2022. Available online: https://data.europa.eu/doi/10.2809/39517 (accessed on 2 October 2023).
- European Food Safety Authority (EFSA); European Centre for Disease Prevention and Control (ECDC). The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2020/2021. EFSA J. 2023, 21, e07867. [Google Scholar] [CrossRef]
- Williams-Nguyen, J.; Sallach, J.B.; Bartelt-Hunt, S.; Boxall, A.B.; Durso, L.M.; McLain, J.E.; Singer, R.S.; Snow, D.D.; Zilles, J.L. Antibiotics and antibiotic resistance in agroecosystems: State of the science. J. Environ. Qual. 2016, 45, 394–406. [Google Scholar] [CrossRef] [PubMed]
- Faleiro, M.L. The mode of antibacterial action of essential oils. In Science against Microbial Pathogens: Communicating Current Research and Technological Advances; Méndez-Vilas, A., Ed.; Formatex Research Center: Badajoz, Spain, 2011; Volume 2, pp. 1143–1156. [Google Scholar]
- Burt, S. Essential oils: Their antibacterial properties and potential applications in foods—A review. Int. J. Food Microbiol. 2004, 94, 223–253. [Google Scholar] [CrossRef]
- Stevanović, Z.D.; Bošnjak-Neumüller, J.; Pajić-Lijaković, I.; Raj, J.; Vasiljević, M. Essential oils as feed additives-future perspectives. Molecules 2018, 23, 1717. [Google Scholar] [CrossRef]
- Mittal, R.P.; Rana, A.; Jaitak, V. Essential oils: An impending substitute of synthetic antimicrobial agents to overcome antimicrobial resistance. Curr. Drug Targets 2019, 20, 605–624. [Google Scholar] [CrossRef]
- Yap, P.S.; Yiap, B.C.; Ping, H.C.; Lim, S.H. Essential oils, a new horizon in combating bacterial antibiotic resistance. Open Microbiol. J. 2014, 8, 6–14. [Google Scholar] [CrossRef]
- Berdejo, D.; Pagán, E.; Merino, N.; Pagán, R.; García-Gonzalo, D. Incubation with a complex orange essential oil leads to evolved mutants with increased resistance and tolerance. Pharmaceuticals 2020, 13, 239. [Google Scholar] [CrossRef]
- Berdejo, D.; Pagán, E.; Merino, N.; García-Gonzalo, D.; Pagán, R. Emerging mutant populations of Listeria monocytogenes EGD-e under selective pressure of Thymbra capitata essential oil question its use in food preservation. Food Res. Int. 2021, 145, 110403. [Google Scholar] [CrossRef]
- Berdejo, D.; Pagán, E.; Merino, N.; Botello-Morte, L.; Pagán, R.; García-Gonzalo, D. Salmonella enterica serovar Typhimurium genetic variants isolated after lethal treatment with Thymbra capitata essential oil (TCO) showed increased resistance to TCO in milk. Int. J. Food Microbiol. 2021, 360, 109443. [Google Scholar] [CrossRef]
- Chueca, B.; Berdejo, D.; Gomes-Neto, N.J.; Pagán, R.; García-Gonzalo, D. Emergence of hyper-resistant Escherichia coli MG1655 derivative strains after applying sub-inhibitory doses of individual constituents of essential oils. Front. Microbiol. 2016, 7, 273. [Google Scholar] [CrossRef]
- Chueca, B.; Renzoni, A.; Berdejo, D.; Pagán, R.; Kelley, W.L.; García-Gonzalo, D. Whole-genome sequencing and genetic analysis reveal novel stress responses to individual constituents of essential oils in Escherichia coli. AEM 2018, 84, e02538-17. [Google Scholar] [CrossRef] [PubMed]
- Berdejo, D.; Chueca, B.; Pagán, E.; Renzoni, A.; Kelley, W.L.; Pagán, R.; Garcia-Gonzalo, D. Sub-inhibitory doses of individual constituents of essential oils can select for Staphylococcus aureus resistant mutants. Molecules 2019, 24, 170. [Google Scholar] [CrossRef]
- Berdejo, D.; Merino, N.; Pagán, E.; García-Gonzalo, D.; Pagán, R. Genetic variants and phenotypic characteristics of Salmonella Typhimurium-resistant mutants after exposure to carvacrol. Microorganisms 2020, 8, 937. [Google Scholar] [CrossRef]
- Li, Y.; Tan, B.; Cen, Z.; Fu, Y.; Zhu, X.; He, H.; Kong, D.; Wu, H. The variation in essential oils composition, phenolic acids and flavonoids is correlated with changes in antioxidant activity during Cinnamomum loureirii bark growth. Arab. J. Chem. 2021, 14, 103249. [Google Scholar] [CrossRef]
- Valková, V.; Ďúranová, H.; Galovičová, L.; Vukovic, N.L.; Vukic, M.; Kowalczewski, P.Ł.; Kačániová, M. Application of three types of cinnamon essential oils as natural antifungal preservatives in wheat bread. Appl. Sci. 2022, 12, 10888. [Google Scholar] [CrossRef]
- Alizadeh Behbahani, B.; Falah, F.; Lavi Arab, F.; Vasiee, M.; Tabatabaee Yazdi, F. Chemical composition and antioxidant, antimicrobial, and antiproliferative activities of Cinnamomum zeylanicum bark essential oil. Evid. Based Complementary Altern. Med. 2020, 2020, 5190603. [Google Scholar] [CrossRef]
- Narayanankutty, A.; Kunnath, K.; Alfarhan, A.; Rajagopal, R.; Ramesh, V. Chemical composition of Cinnamomum verum leaf and flower essential oils and analysis of their antibacterial, insecticidal, and larvicidal properties. Molecules 2021, 26, 6303. [Google Scholar] [CrossRef]
- Mancini, E.; Senatore, F.; Del Monte, D.; De Martino, L.; Grulova, D.; Scognamiglio, M.; Snoussi, M.; De Feo, V. Studies on chemical composition, antimicrobial and antioxidant activities of five Thymus vulgaris L. essential oils. Molecules 2015, 20, 12016–12028. [Google Scholar] [CrossRef]
- Merino, N.; Berdejo, D.; Bento, R.; Salman, H.; Lanz, M.; Maggi, F.; Sánchez-Gómez, S.; García-Gonzalo, D.; Pagán, R. Antimicrobial efficacy of Thymbra capitata (L.) Cav. essential oil loaded in self-assembled zein nanoparticles in combination with heat. Ind. Crop. Prod. 2019, 133, 98–104. [Google Scholar] [CrossRef]
- van Den Dool, H.; Kratz, P.D. A generalization of the retention index system including linear temperature programmed gas—Liquid partition chromatography. J. Chromatogr. A 1963, 11, 463–471. [Google Scholar] [CrossRef]
- Adams, R. Identification of Essential Oil Components by Gas Chromatography/Quadrupole Mass Spectroscopy; Allured Publishing Corporation: Carol Stream, IL, USA, 2005; pp. 65–120. [Google Scholar]
- NIST. Mass Spectral Library (NIST/EPA/NIH); NIST: Gaithersburg, MD, USA, 2017.
- Mondello, L. Flavors and Fragrances of Natural and Synthetic Compounds; Wiley: Hoboken, NJ, USA, 2015. [Google Scholar]
- Manu, D.; Mendonca, A.F.; Daraba, A.; Dickson, J.S.; Sebranek, J.; Shaw, A.; Wang, F.; White, S. Antimicrobial efficacy of cinnamaldehyde against Escherichia coli O157:H7 and Salmonella enterica in carrot juice and mixed berry juice held at 4 °C and 12 °C. Foodborne Pathog. Dis. 2017, 14, 302–307. [Google Scholar] [CrossRef] [PubMed]
- García-Salinas, S.; Elizondo-Castillo, H.; Arruebo, M.; Mendoza, G.; Irusta, S. Evaluation of the antimicrobial activity and cytotoxicity of different components of natural origin present in essential oils. Molecules 2018, 23, 1399. [Google Scholar] [CrossRef]
- Devi, K.P.; Nisha, S.A.; Sakthivel, R.; Pandian, S.K. Eugenol (an essential oil of clove) acts as an antibacterial agent against Salmonella typhi by disrupting the cellular membrane. J. Ethnopharmacol. 2010, 130, 107–115. [Google Scholar] [CrossRef] [PubMed]
- Engel, J.B.; Heckler, C.; Tondo, E.C.; Daroit, D.J.; da Silva Malheiros, P. Antimicrobial activity of free and liposome-encapsulated thymol and carvacrol against Salmonella and Staphylococcus aureus adhered to stainless steel. Int. J. Food Microbiol. 2017, 252, 18–23. [Google Scholar] [CrossRef]
- Sepahvand, S.; Amiri, S.; Radi, M.; Akhavan, H.-R. Antimicrobial activity of thymol and thymol-nanoemulsion against three food-borne pathogens inoculated in a sausage model. Food Bioprocess Technol. 2021, 14, 1936–1945. [Google Scholar] [CrossRef]
- Jianu, C.; Pop, G.; Lukinich-Gruia, A.; Horhat, F. Chemical composition and antimicrobial activity of essential oils of lavender (Lavandula angustifolia) and lavandin (Lavandula x intermedia) grown in western Romania. Int. J. Agric. Biol. 2013, 15, 772–776. [Google Scholar]
- EUCAST. MIC and Zone Diameter Distributions and ECOFFs. Available online: http://www.eucast.org (accessed on 13 January 2023).
- Ait-Ouazzou, A.; Cherrat, L.; Espina, L.; Lorán, S.; Rota, C. The antimicrobial activity of hydrophobic essential oil constituents acting alone or in combined processes of food preservation. Innov. Food Sci. Emerg. Technol. 2011, 12, 320–329. [Google Scholar] [CrossRef]
- Dafale, N.A.; Semwal, U.P.; Rajput, R.K.; Singh, G.N. Selection of appropriate analytical tools to determine the potency and bioactivity of antibiotics and antibiotic resistance. J. Pharm. Anal. 2016, 6, 207–213. [Google Scholar] [CrossRef]
- Thi, T.D.; López, E.; Rodríguez-Rojas, A.; Rodríguez-Beltrán, J.; Couce, A.; Guelfo, J.R.; Castañeda-García, A.; Blázquez, J. Effect of recA inactivation on mutagenesis of Escherichia coli exposed to sublethal concentrations of antimicrobials. J. Antimicrob. Chemother. 2011, 66, 531–538. [Google Scholar] [CrossRef]
- Jara, L.M.; Cortés, P.; Bou, G.; Barbé, J.; Aranda, J. Differential roles of antimicrobials in the acquisition of drug resistance through activation of the SOS response in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2015, 59, 4318–4320. [Google Scholar] [CrossRef]
- Tzeng, Y.L.; Berman, Z.; Toh, E.; Bazan, J.A.; Turner, A.N.; Retchless, A.C.; Wang, X.; Nelson, D.E.; Stephens, D.S. Heteroresistance to the model antimicrobial peptide polymyxin B in the emerging Neisseria meningitidis lineage 11.2 urethritis clade: Mutations in the pilMNOPQ operon. Mol. Microbiol. 2019, 111, 254–268. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Rojas, A.; Makarova, O.; Müller, U.; Rolff, J. Cationic peptides facilitate iron-induced mutagenesis in bacteria. PLoS Genet. 2015, 11, e1005546. [Google Scholar] [CrossRef] [PubMed]
- Kohanski, M.A.; DePristo, M.A.; Collins, J.J. Sublethal antibiotic treatment leads to multidrug resistance via radical-induced mutagenesis. Mol. Cell 2010, 37, 311–320. [Google Scholar] [CrossRef] [PubMed]
- Hriouech, S.; Akhmouch, A.A.; Tanghort, M.; Chefchaou, H.; Mzabi, A.; Chami, N.; Remmal, A. In vitro and in vivo comparison of changes in antibiotics susceptibility of E. coli and chicken’s intestinal flora after exposure to amoxicillin or thymol. Vet. Med. Int. 2020, 2020, 8824008. [Google Scholar] [CrossRef]
- Lee, C.; Kim, I.; Lee, J.; Lee, K.L.; Min, B.; Park, C. Transcriptional activation of the aldehyde reductase YqhD by YqhC and its implication in glyoxal metabolism of Escherichia coli K-12. J. Bacteriol. 2010, 192, 4205–4214. [Google Scholar] [CrossRef]
- Pérez, J.M.; Arenas, F.A.; Pradenas, G.A.; Sandoval, J.M.; Vásquez, C.C. Escherichia coli YqhD exhibits aldehyde reductase activity and protects from the harmful effect of lipid peroxidation-derived aldehydes. J. Biol. Chem. 2008, 283, 7346–7353. [Google Scholar] [CrossRef]
- Merchel Piovesan Pereira, B.; Adil Salim, M.; Rai, N.; Tagkopoulos, I. Tolerance to glutaraldehyde in Escherichia coli mediated by overexpression of the aldehyde reductase YqhD by YqhC. Front. Microbiol. 2021, 12, 680553. [Google Scholar] [CrossRef]
- Visvalingam, J.; Hernandez-Doria, J.D.; Holley, R.A. Examination of the genome-wide transcriptional response of Escherichia coli O157:H7 to cinnamaldehyde exposure. AEM 2013, 79, 942–950. [Google Scholar] [CrossRef]
- Yuan, W.; Seng, Z.J.; Kohli, G.S.; Yang, L.; Yuk, H.-G. Stress resistance development and genome-wide transcriptional response of Escherichia coli O157: H7 adapted to sublethal thymol, carvacrol, and trans-cinnamaldehyde. AEM 2018, 84, 01616–01618. [Google Scholar] [CrossRef]
- Chueca, B.; Pagán, R.; García-Gonzalo, D. Oxygenated monoterpenes citral and carvacrol cause oxidative damage in Escherichia coli without the involvement of tricarboxylic acid cycle and Fenton reaction. Int. J. Food Microbiol. 2014, 189, 126–131. [Google Scholar] [CrossRef]
- Lyu, Z.; Yang, A.; Villanueva, P.; Singh, A.; Ling, J. Heterogeneous flagellar expression in single Salmonella cells promotes diversity in antibiotic tolerance. mBio 2021, 12, 02374-21. [Google Scholar] [CrossRef] [PubMed]
- Tirumalai, M.R.; Karouia, F.; Tran, Q.; Stepanov, V.G.; Bruce, R.J.; Ott, C.M.; Pierson, D.L.; Fox, G.E. Evaluation of acquired antibiotic resistance in Escherichia coli exposed to long-term low-shear modeled microgravity and background antibiotic exposure. mBio 2019, 10, 02637-18. [Google Scholar] [CrossRef] [PubMed]
- Forbes, S.; Morgan, N.; Humphreys, G.J.; Amézquita, A.; Mistry, H.; McBain, A.J. Loss of function in Escherichia coli exposed to environmentally relevant concentrations of benzalkonium chloride. AEM 2019, 85, 02417–02418. [Google Scholar] [CrossRef]
- Scheu, P.D.; Witan, J.; Rauschmeier, M.; Graf, S.; Liao, Y.F.; Ebert-Jung, A.; Basché, T.; Erker, W.; Unden, G. CitA/CitB two-component system regulating citrate fermentation in Escherichia coli and its relation to the DcuS/DcuR system in vivo. J. Bacteriol. 2012, 194, 636–645. [Google Scholar] [CrossRef] [PubMed]
- Miller, C.; Ingmer, H.; Thomsen, L.E.; Skarstad, K.; Cohen, S.N. DpiA binding to the replication origin of Escherichia coli plasmids and chromosomes destabilizes plasmid inheritance and induces the bacterial SOS response. J. Bacteriol. 2003, 185, 6025–6031. [Google Scholar] [CrossRef]
- Rycovska-Blume, A.; Lü, W.; Andrade, S.; Fendler, K.; Einsle, O. Structural and functional studies of NirC from Salmonella typhimurium. Methods Enzymol. 2015, 556, 475–497. [Google Scholar] [CrossRef]
- Adamowicz, E.M.; Muza, M.; Chacón, J.M.; Harcombe, W.R. Cross-feeding modulates the rate and mechanism of antibiotic resistance evolution in a model microbial community of Escherichia coli and Salmonella enterica. PLoS Pathog. 2020, 16, e1008700. [Google Scholar] [CrossRef]
- Sun, S.; Selmer, M.; Andersson, D.I. Resistance to β-lactam antibiotics conferred by point mutations in penicillin-binding proteins PBP3, PBP4 and PBP6 in Salmonella enterica. PLoS ONE 2014, 9, e97202. [Google Scholar] [CrossRef]
- Olaitan, A.O.; Morand, S.; Rolain, J.-M. Mechanisms of polymyxin resistance: Acquired and intrinsic resistance in bacteria. Front. Microbiol. 2014, 5, 643. [Google Scholar] [CrossRef]
- Lee, H.; Hsu, F.F.; Turk, J.; Groisman, E.A. The PmrA-regulated pmrC gene mediates phosphoethanolamine modification of lipid A and polymyxin resistance in Salmonella enterica. J. Bacteriol. 2004, 186, 4124–4133. [Google Scholar] [CrossRef]
- Zhou, Z.; Ribeiro, A.A.; Lin, S.; Cotter, R.J.; Miller, S.I.; Raetz, C.R. Lipid A modifications in polymyxin-resistant Salmonella typhimurium: PMRA-dependent 4-amino-4-deoxy-L-arabinose, and phosphoethanolamine incorporation. J. Biol. Chem. 2001, 276, 43111–43121. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Negrea, A.; Rhen, M.; Andersson, D.I. Genetic analysis of colistin resistance in Salmonella enterica serovar Typhimurium. Antimicrob. Agents Chemother. 2009, 53, 2298–2305. [Google Scholar] [CrossRef] [PubMed]
- Ghasemian, A.; Shafiei, M.; Hasanvand, F.; Shokouhi Mostafavi, S.K. Carbapenem and colistin resistance in Enterobacteriaceae: Worldwide spread and future perspectives. Rev. Med. Microbiol. 2018, 29, 173–176. [Google Scholar] [CrossRef]
- Mahalakshmi, S.; Sunayana, M.R.; SaiSree, L.; Reddy, M. yciM is an essential gene required for regulation of lipopolysaccharide synthesis in Escherichia coli. Mol. Microbiol. 2014, 91, 145–157. [Google Scholar] [CrossRef]
- Yan, L.; Dong, H.; Li, H.; Liu, X.; Deng, Z.; Dong, C.; Zhang, Z. Uncovering lipopolysaccharide regulation in bacteria via the critical lipid binding tunnel of YciS/YciM. iScience 2022, 25, 104988. [Google Scholar] [CrossRef]
- Lallemand, E.A.; Lacroix, M.Z.; Toutain, P.-L.; Boullier, S.; Ferran, A.A.; Bousquet-Melou, A. In vitro degradation of antimicrobials during use of broth microdilution method can increase the measured minimal inhibitory and minimal bactericidal concentrations. Front. Microbiol. 2016, 7, 2051. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically, 9th ed.; CLSI Document M07-A9; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2012; ISBN 1-56238-783-9 (Print), ISBN 1-56238-784-7 (Electronic). [Google Scholar]
- Friedman, M.; Henika, P.R.; Mandrell, R.E. Bactericidal activities of plant essential oils and some of their isolated constituents against Campylobacter jejuni, Escherichia coli, Listeria monocytogenes, and Salmonella enterica. J. Food Prot. 2002, 65, 1545–1560. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, A.; Laureti, L.; Crussard, S.; Abida, H.; Rodríguez-Rojas, A.; Blázquez, J.; Baharoglu, Z.; Mazel, D.; Darfeuille, F.; Vogel, J.; et al. β-Lactam antibiotics promote bacterial mutagenesis via an RpoS-mediated reduction in replication fidelity. Nat. Commun. 2013, 4, 1610. [Google Scholar] [CrossRef]
- Rosche, W.A.; Foster, P.L. Determining mutation rates in bacterial populations. Methods 2000, 20, 4–17. [Google Scholar] [CrossRef]
- Andersson, D.I.; Hughes, D. Microbiological effects of sublethal levels of antibiotics. Nat. Rev. Microbiol. 2014, 12, 465–478. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Wallis, J.W.; McLellan, M.D.; Larson, D.E.; Kalicki, J.M.; Pohl, C.S.; McGrath, S.D.; Wendl, M.C.; Zhang, Q.; Locke, D.P.; et al. BreakDancer: An algorithm for high-resolution mapping of genomic structural variation. Nat. Methods 2009, 6, 677–681. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010, 38, 164. [Google Scholar] [CrossRef]



| AEN | COLIFIT | |||||
|---|---|---|---|---|---|---|
| No | Component a | RI b | RI Lit c | % d | % | ID e |
| 1 | α-pinene | 933 | 932 | - | 0.17 ± 0.0 | Std,RI,MS |
| 2 | camphene | 948 | 946 | - | 0.03 ± 0.0 | Std,RI,MS |
| 3 | benzaldehyde | 959 | 952 | - | 0.05 ± 0.0 | Std,RI,MS |
| 4 | α-terpinene | 1017 | 1014 | - | 0.03 ± 0.0 | Std,RI,MS |
| 5 | p-cymene | 1025 | 1020 | - | 1.30 ± 0.0 | Std,RI,MS |
| 6 | limonene | 1029 | 1024 | - | 0.37 ± 0.0 | Std,RI,MS |
| 7 | γ-terpinene | 1059 | 1054 | - | 0.72 ± 0.0 | Std,RI,MS |
| 8 | diallyl disulphide | 1078 | 1079 | - | 0.13 ± 0.0 | RI,MS |
| 9 | linalool | 1101 | 1095 | - | 0.05 ± 0.0 | Std,RI,MS |
| 10 | citronellal | 1155 | 1148 | - | 1.98 ± 0.0 | Std,RI,MS |
| 11 | terpinen-4-ol | 1177 | 1174 | - | 0.06 ± 0.0 | Std,RI,MS |
| 12 | (Z)-cinnamaldehyde | 1219 | 1217 | - | 0.10 ± 0.0 | Std,RI,MS |
| 13 | citronellol | 1230 | 1223 | - | 0.18 ± 0.0 | Std,RI,MS |
| 14 | neral | 1242 | 1235 | - | 1.93 ± 0.0 | Std,RI,MS |
| 15 | geraniol | 1256 | 1249 | - | 0.90 ± 0.0 | Std,RI,MS |
| 16 | (E)-cinnamaldehyde | 1270 | 1267 | 87.12 ± 0.2 | 43.93 ± 0.7 | Std,RI,MS |
| 17 | geranial | 1272 | 1264 | - | 3.12 ± 0.7 | Std,RI,MS |
| 18 | thymol | 1293 | 1289 | - | 29.83 ± 0.0 | Std,RI,MS |
| 19 | carvacrol | 1302 | 1298 | - | 10.56 ± 0.1 | Std,RI,MS |
| 20 | citronellyl acetate | 1356 | 1350 | - | 0.03 ± 0.0 | RI,MS |
| 21 | eugenol | 1358 | 1356 | 10.83 ± 0.2 | 2.99 ± 0.2 | Std,RI,MS |
| 22 | α-copaene | 1377 | 1374 | - | 0.04 ± 0.0 | Std,RI,MS |
| 23 | geranyl acetate | 1386 | 1379 | - | 0.24 ± 0.0 | RI,MS |
| 24 | (E)-caryophyllene | 1421 | 1417 | 1.40 ± 0.0 | 0.96 ± 0.0 | Std,RI,MS |
| 25 | coumarin | 1435 | 1432 | - | 0.05 ± 0.0 | RI,MS |
| 26 | δ-cadinene | 1526 | 1522 | - | 0.09 ± 0.0 | RI,MS |
| Total identified | 99.35 ± 0.0 | 99.83 ± 0.0 | ||||
| Aldehydes | - | 5.10 ± 0.7 | ||||
| Monoterpene hydrocarbons | - | 1.32 ± 0.0 | ||||
| Monoterpenoids | - | 43.54 ± 0.2 | ||||
| Phenylpropanoids | 97.96 | 47.02 ± 0.5 | ||||
| Sesquiterpene hydrocarbons | 1.40 | 1.08 ± 0.0 | ||||
| Others | - | 1.76 ± 0.0 |
| Bacterial Strain | AEN | COLIFIT | Amoxicillin | Colistin | ||||
|---|---|---|---|---|---|---|---|---|
| MIC | MBC | MIC | MBC | MIC | MBC | MIC | MBC | |
| EcWT | 200 | 400 | 350 | 450 | 8 | 8 | 1 | 1 |
| SeWT | 150 | 350 | 350 | 350 | 1 | 1 | 2 | 4 |
| Bacterial Strain | AEN | COLIFIT | Amoxicillin | Colistin | Thymol | Cinnamaldehyde |
|---|---|---|---|---|---|---|
| EcWT | 200 | 350 | 8 | 1 | 300 | 200 |
| EcAEN10 | 250 | 400 | 8 | 1 | 300 | 250 |
| EcAEN20 | 250 | 450 | 8 | 1 | 300 | 250 |
| EcCOLIFIT10 | 250 | 400 | 8 | 1 | 300 | 250 |
| EcCOLIFIT20 | 250 | 450 | 8 | 1 | 300 | 250 |
| EcAmox20 | 200 | 400 | 16 | 1 | 300 | 250 |
| Bacterial Strain | AEN | COLIFIT | Amoxicillin | Colistin | Thymol | Cinnamaldehyde |
|---|---|---|---|---|---|---|
| SeWT | 150 | 350 | 1 | 2 | 250 | 150 |
| SeAmox10 | nd | nd | 2 | nd | nd | nd |
| SeAmox20 | 150 | 350 | 2 | 2 | 250 | 150 |
| SeCol10 | nd | nd | nd | 8 | nd | nd |
| SeCol20 | 150 | 250 | 1 | 8 | 250 | 150 |
| Strain | Genetic Modification | Position | Gene |
|---|---|---|---|
| EcAEN10 | 22-bp deletion | 3,154,511–3,154,531 | yqhC |
| EcAEN20 | 5-kb deletion | 1,973,201–1,978,600 | cheW, cheA, motB, motA, motR, flhC, flhD |
| EcCOLIFIT10 | SNV | 3,156,163 G809A Gly270Asp | yqhD |
| 5-kb deletion | 1,973,663–1,978,501 | cheA, motB, motA, motR, flhC, flhD | |
| EcCOLIFIT20 | SNV | 3,156,163 G809A Gly270Asp | yqhD |
| 5-kb deletion | 1,973,663–1,978,501 | cheA, motB, motA, motR, flhC, flhD | |
| EcAmox20 | Deletion | 652,049 | Intergenic region (DpiA binding site) |
| Strain | Genetic Modification | Position | Gene |
|---|---|---|---|
| SeAmox10 | SNV A1988T Gln663Leu | 143,332 | ftsI |
| Insertion 107 | 643,920 | fepA | |
| SNV T215C Val72Ala | 3,626,869 | nirC | |
| SeAmox20 | SNV A1988T Gln663Leu | 143,332 | ftsI |
| Insertion 107 | 643,920 | fepA | |
| SNV T215C Val72Ala | 3,626,869 | nirC | |
| SeCol10 | Insertion 107 | 643,922 | fepA |
| SNV A499C Thr167Pro | 695,332 | lipA | |
| SNV T215C Val72Ala | 3,626,869 | nirC | |
| SNV A274C Thr92Pro | 4,533,452 | basS | |
| SeCol20 | Insertion 107 | 643,922 | fepA |
| SNV T128G Val43Gly | 1,802,551 | yciM | |
| SNV T215C Val72Ala | 3,626,869 | nirC | |
| SNV A274C Thr92Pro | 4.533.452 | basS |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Merino, N.; Berdejo, D.; Pagán, E.; Girard, C.; Kerros, S.; Spinozzi, E.; Pagán, R.; García-Gonzalo, D. Phenotypic and Genotypic Comparison of Antimicrobial-Resistant Variants of Escherichia coli and Salmonella Typhimurium Isolated from Evolution Assays with Antibiotics or Commercial Products Based on Essential Oils. Pharmaceuticals 2023, 16, 1443. https://0-doi-org.brum.beds.ac.uk/10.3390/ph16101443
Merino N, Berdejo D, Pagán E, Girard C, Kerros S, Spinozzi E, Pagán R, García-Gonzalo D. Phenotypic and Genotypic Comparison of Antimicrobial-Resistant Variants of Escherichia coli and Salmonella Typhimurium Isolated from Evolution Assays with Antibiotics or Commercial Products Based on Essential Oils. Pharmaceuticals. 2023; 16(10):1443. https://0-doi-org.brum.beds.ac.uk/10.3390/ph16101443
Chicago/Turabian StyleMerino, Natalia, Daniel Berdejo, Elisa Pagán, Claire Girard, Sylvain Kerros, Eleonora Spinozzi, Rafael Pagán, and Diego García-Gonzalo. 2023. "Phenotypic and Genotypic Comparison of Antimicrobial-Resistant Variants of Escherichia coli and Salmonella Typhimurium Isolated from Evolution Assays with Antibiotics or Commercial Products Based on Essential Oils" Pharmaceuticals 16, no. 10: 1443. https://0-doi-org.brum.beds.ac.uk/10.3390/ph16101443