1. Introduction
Azo dyes account for 70% of the 9.9 million tons of industrial dye used annually, with a global turnover valued at USD 30.42 billion [
1,
2]. The continued demand for dyes and pigments causes an increase in the supply rate of 3.5% per annum. Most of the dyes synthesised contain azo compounds and are predominantly used in textile, paper, food, printing, cosmetic and leather industries [
3]. These azo dyes are extensively used in fabric manufacturing due to low cost, ease of preparation, fastness, versatility and intensity of the colours [
4]. Certain azo dyes contain chemical groups, which have a high affinity for metal ions [
5]. These enhanced properties provide a high degree of chemical, biological and photocatalytic stability. Amongst azo dyes, Reactive Red 120 (RR120) is one of the widely used dyes in the textile industry due to it providing a high degree of chemical, biological and photocatalytic stabilities [
1]. Being a diazo, RR120 is one of the hardest and most durable of reactive dyes and can resist degradation. Nevertheless, their resistance to breakdown due to time, exposure to sunlight, detergents, water and microorganisms results in poor degradation in the environment [
6].
Pollution and poisoning by azo dyes still happen to this day. With an ever-increasing number of cancers, dye plays important role in contributing as a breakdown product producing toxic amines and benzene [
7]. These toxic metabolics easily get into us through consuming water and fish exposed to dye waste. Discharge of untreated textile waste into nearby streams and river can cause anoxic conditions that are lethal to aquatic organisms [
8]. Therefore, chemical treatment of the effluents is often employed to treat the waste. Chemical processes, such as the oxidative process, Fenton’s reagent, ozonation, photochemical, cucurbituril and electrochemical destruction, are examples used for decolourisation purposes [
9]. This method is highly effective, faster, and most importantly can be done on large scale; but the drawback of this mechanism is quite distressing [
9]. Some of these processes are dye-specific, making mix dye eluents take several processes to completely decolourise them. Moreover, the application of such technologies is usually applied by high-end industrial producers and is limited to small scale manufacturers due to the cost of handling and maintenance. Therefore, the removal of azo dyes requires an alternative cheap and environmentally friendly approach whereby the role of bacteria could provide such.
The use of microorganisms in treating dye wastes is one of the most favourable processes in comparison to other applications due to its practicality, productivity, simplicity, and inexpensiveness [
10]. Bioremediation allows the bacteria to consume the available dye compound during cell propagation. Unlike biosorption, even though the dyes have been removed from the wastewater, the absorbate still holds the dye compounds which requires further physio-chemical treatment to reduce it, whereby decolourisation using bacteria solves both problems by eliminating the dye compound and reducing the need for chemical exposure [
11]. Various azo dye decolourisation using bacteria, such as Reactive Black 5 [
12], Black 5 [
13], Methyl Orange [
14], Acid Red [
15], and Green 19 [
16], have been reported before.
For over a century, microbial decolourisation of azo dyes has been an unsolved puzzle among scientists. Previously, in microbial azo dye decolourisation, the focus was centred on isolating bacteria or consortia that have a higher azo dye tolerance level needed as a tool for bioremediation [
17,
18]. To achieve this, various additional carbon sources and a higher concentration of co-substrate were introduced. During the last ten years, the attention has shifted towards isolating microorganisms with the ability to decolourise azo dyes with complete mineralisation ability [
14]. The focus changed mainly because degraded metabolites of azo dye were found to be toxic [
19].
Improper cleavage of azo dyes during the decolourisation process could result in toxic metabolites that are far more toxic than the parent compound. As such, these compounds generally consist of benzene and aniline groups, which are a precursor to carcinogenic and mutagenic effects [
19]. Even though decolourisation has occurred, there is always a chance that colourless metabolites produced by the bacteria end up being toxic. A race to find the best strains to solve azo dye pollution ended up manifesting more problems rather than providing a good solution. This dilemma has been going on for the past few decades, leading to various methods and media used for the purpose of isolation. However, the role of method and its influence on media used for azo dye decolourisation has not been compared nor studied before. Having this understating could provide crucial data needed to better isolate azo dye consuming bacteria. Since each media composition is different to the others, the role of media in supporting azo dye decolourisation needs to be elucidated.
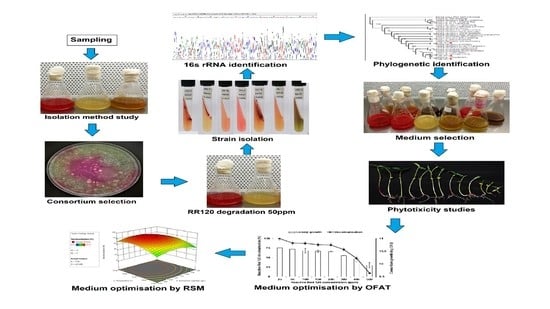
In this study, dye contaminated samples from the heavily polluted industrial waste site were obtained from Juru River in Malaysia. These samples were examined using a different method of isolation to identify bacterial strains able to decolourise RR120 as sole carbon source. The role of different media compositions and co-substrates in sustaining maximum RR120 decolourisation was also further investigated. The toxic level of the produced metabolites was assessed using mung beans. Since decolourisation of azo dye involving diazo as a sole carbon source is a slow process, factors affecting decolourisation were optimised to reinforce the process of degradation. Further optimisation was done using a surface methodology approach (RSM) which utilised a mathematical and statistical approach to develop and study the interaction between two factors. To the best of our knowledge, this is the first isolation of aerobic decolourisation of RR120 by a novel bacterial consortium.
2. Materials and Methods
2.1. Chemicals, Reagents, and Equipment
The Reactive Red 120 (RR120) used in this research was purchased from Sigma, Aldrich, CO, USA. Nutrient agar, yeast extracts and nutrient broth were obtained from Friedemann Schmidt Shd. Bhd. Malaysia. Tris and acetate buffers were obtained from Merck KGaA, Germany. Meanwhile, other chemicals used in this study were obtained from Fisher Malaysia.
2.2. Sample Soil and Water
Juru River was chosen as a prime location for sampling as the state contributed 28.2% of total water pollution originating from the textile industry [
20]. Most of these industrial factories are located near to Juru Riverside. Industrial textile wastewater effluent was collected in a 20 mL sterilised falcon tube and samples were brought to the lab and processed within 24 h. For sample preparation, 10 uL cycloheximide was introduced to prevent fungal growth. The locations of samples collected were recorded using the map coordinates provided by Google Earth to locate the exact locations from which the samples were collected (
Table 1)
2.3. Screening for RR120 Dye Decolourising Bacteria
The study was initiated by studying the best for in obtaining bacteria capable of decolourising Reactive Red 120 as a sole carbon source.
2.3.1. Method 1: Isolation Using Nutrient Broth
Method number 1 is commonly used to rapidly isolate dye decolourising bacteria. This involves samples inoculated in nutrient broth containing some sort of azo dye [
21,
22,
23]. Briefly, a 1 mL of sample was inoculated in 100 mL nutrient broth containing 100 ppm filter-sterilised RR120 dye. The culture was incubated in a 250 mL conical flask at room temperature under static conditions for 72. After 72 h of incubation, the sample was streaked in nutrient agar containing 100 ppm RR120. The morphologically distinct bacterial isolates showing a clear zone of decolourisation were selected for identification.
2.3.2. Method 2: Isolation through a Straightforward Process
Method 2 is a straightforward method for isolating dye decolourising bacteria without the process of acclimatisation. This was done by inoculating 1 mL sample in minimal salt medium (MSM) containing 100 ppm RR120 [
24,
25]. The composition used MSM in g/L of dH
2O was pH (7.0 to 7.2) yeast extract (1.0) Na
2HPO
4 (1.0), KH
2PO
4 (1.0), NaCl (1.0), MgSO
4.7H
2O (0.5), CaCl
2 (0.01) [
26]. A 1 mL sample was incubated in 100 mL sterilised MSM in a 250 mL conical flask containing 100 ppm filter-sterilised RR120. The culture medium was incubated for 72 h in shaking condition at 31 °C. The dye decolourising strain was isolated using streaking on MSM agar containing 100 ppm RR120, which showed a clear zone of decolourisation.
2.3.3. Method 3: Isolation through Long Acclimatisation Process
Method 3 involves the acclimation of bacterial strains to improve decolourisation capacity by slowly introducing an increased concentration of RR120 while reducing substrate and carbon availability [
27,
28]. A 1 mL sample was first inoculated in nutrient broth containing 25 ppm RR120 and was incubated at 31 °C for 72 h in shaking conditions (150 rpm). A 1 mL of sample was taken from a previously grown culture and incubated in another 100 mL of MSM-containing 8 g/L glucose, 3 g/L yeast extract and 25 ppm RR120. The inoculated medium was incubated at 31 °C under shaking conditions (150 rpm) for a week. From this suspension, 1 mL was transferred into fresh MSM-containing 5 g/L glucose and 3 g/L yeast extract with 50 ppm RR120. A similar successive transfer was done with 2 g/L glucose, 3 g/L yeast extract and 100 ppm RR120. The next successive transfer was done with 3 g/L yeast extract and 150 ppm RR120 followed by 1 g/L yeast extract and 200 ppm RR120. The final successive transfer was done with MSM containing 200 ppm RR120 only. For each successive, the culture medium was incubated at 31 °C for a week in shaking conditions. Those bacterial strains that grew on MSM (no glucose and yeast extract added) containing 200 ppm RR120 as the sole carbon source was selected for further identification.
2.4. Analytical Method
Decolourisation of Reactive Red 120 was determined by colourimetric determination. The absorbance of each solution was determined using a 525 nm wavelength against blank using a Shimadzu U.V. Mini 1240 spectrophotometer. Meanwhile, bacterial growth was measured using a colony-forming unit per millilitre technique (CFU/mL) and optical density (OD) at 600 nm [
21].
2.5. Identification of Isolates
The biochemical test was done to determine the bacteria species based on physiological differences. The included test was Gram staining, oxidase/catalase test, indole and nitrate production, citrate, Voges-Proskauer, urease and sugar test. The genomic DNA of the selected strains were extracted with the innuPREP extraction kit (Analytik Jena GmbH, Jena, Germany), following the manufacturer’s recommended procedure. The Polymerase chain reaction (PCR) product was produced using a Biometra T-Gradient Thermocycler. The mixture contains 1 µL Template DNA, 14.2 µL dH2O, 1.2 µL of 25 mM MgCl2, 2 µL of 10 × Taq polymerase buffer, 0.5 µL of 10 mM Deoxynucleotide triphosphate mix, 0.5 µL of 10 µM forward primer, 0.5 µL of 10 µM reverse primer, and 0.1 µL Taq DNA polymerase. PCR universal primer; 27F: 5′-AGA GTT TGA TCC TGG CTC AG-3′ and 1492R: 5′-TAC GGT TAC CTT GTT ACG ACT T-3′, corresponding to forward and reverse primers of 16S rRNA, was used to amplify the 16S rRNA gene of the isolates. The thermal cycler protocol was comprised of an initial 4 min denaturation at 95 °C for 1 cycle, followed by 30 cycles of 1 min denaturation at 95 °C, primer hybridisation at 52 °C for 1 min and elongation at 72 °C for 1 min with 1 final cycle for a 7 min extension step at 72 °C. Successful PCR product was purified using a GeneJET Gel Extraction and DNA Cleanup Kit (Thermo Scientific, Waltham, MA, USA) before a sequencing process using an ABI 3730xl DNA Analyzer (Applied Biosystems, Foster City, CA, USA).
2.6. Phylogenetic Analysis
The top 20 16s rRNA sequence with the highest similarity of the related isolate’s sequence was obtained from GenBank using BLAST (Basic Local Alignment Search Tools) (
www.ncbi.nlm.nhi.gov/BLAST/bl2seq/, accessed on 10 August 2020). Using Clustal Omega program (
www.ebi.ac.uk/Tools/msa/clustalo/, accessed on 10 August 2020), the 20 sequence from the GenBank was aligned with a target isolate’s gene. The obtained aligned files were analysed using evolutionary analysis software PHYLIP v 3.6. The neighbour-joining method was used to infer evolutionary history [
29].
Bacillus cereus ATCC 14,579 (Accession no. MG708176) was used as an out-group to construct the phylogenetic tree.
2.7. Media Selection for Optimal RR120 Decolourisation
The effect of media composition on the obtained consortium for RR120 decolourisation was investigated. Five media with different composition was used for the best media selection (
Table 2). Each consortium was inoculated in MSM containing filter-sterilised 50 ppm RR120 and incubated at room temperature in shaking conditions (150 rpm) at 31 °C. The shaking condition was done regardless of the original media condition to prevent anaerobic decolourisation. Resting cells were used to remove any nutrient residue from previously cultured medium that could influence dye decolourisation. Reading of dye decolourisation was taken at 24 h.
2.8. Phytotoxicity Study Using Vigna Radiata
The phytotoxicity study was carried out using mung bean (
Vigna radiata). All the mung bean seeds were first sterilised using 70% ethanol for 3 min followed by 5% sodium hypochlorite for another 3 min [
35]. Then those sterilised mung beans were washed 5 times with sterilised dH
2O and soaked for 3 min in sterilised dH
2O [
36]. Decolourised culture medium on the 24 h was centrifuged at 10,000×
g for 30 min at 4 °C and the resulting supernatant was filter-sterilised using a 0.22 μm pore filter. The sample was given 3 mL of filter sterilised untreated/treated sample per day. The control set was carried out using distilled water at the same time. The germination (%) and length of shoot and root was recorded after day 7.
2.9. Effect of Co-Substrate on Decolourisation of RR120
The role of co-substrate in improving the decolourisation activity of the obtained consortium was investigated in this study. The purpose of the experiment was to determine the ability of said bacteria to decolourise RR120 without any available co-substrate. This was done by removing any glucose and yeast extract from the media composition based on
Table 2.
2.10. Effect of Yeast Extract on RR120 Decolourisation
Decolourisation without any presence of co-substrate is a relatively slow process and rare. Therefore, to enhance the decolourisation azo dye, the yeast extract is supplemented as a co-substrate. The yeast concentration of 0 g/L to 5 g/L with an interval of 0.5 g/L was supplemented into 100 mL of the best MSM in sustaining decolourisation of RR120 as the sole carbon source.
2.11. Optimisation of RR120 Decolourisation Using One-Factor-At-A-Time
The best medium in sustaining in maximum RR120 decolourisation based on
Table 2 and its condition was further optimised in this study. Four different parameters, such as nitrogen source and concentration, pH, temperature, and dye concentration, were used to study the growth and decolourisation rates. Five different nitrogen sources were used in the experiment—0.5 g/L each of ammonia chloride (NH
4Cl), ammonium sulphate (NH
4SO
4), potassium nitrate (KNO
3), urea (CH
4N
2O) and magnesium nitrate (Mg(NO
3)
2). A pH ranging from 5 to 9 was optimised using appropriate buffers. The varied pH was adjusted using different overlapping buffers systems whereby at pH 5.0 and 6.0, 50 mM a citric buffer was used, while for pH 6.0, 7.0 and 8.0, 50 mM, phosphate buffer was utilised. For pH 8.0 and 9.0, 50 mM, Tris buffer was used. To study the effect of temperature on RR120 decolourisation, temperature ranging from 20 to 50 °C were used. The ability of bacteria to degrade a high concentration of RR120 was studied to ascertain the optimum concentration of RR120. RR120 concentrations of 25, 50, 100, 150, 200, 300, 400, and 500 ppm.
2.12. Optimisation of RR120 Decolourisation Using Response Surface Methodology
Statistical optimisation using RSM was carried out based on one-factor-at-a-time (OFAT) factors range levels. A Design Expert v 13.0 (Trial Version, Stat- Ease Inc., Minneapolis, MN, USA) was utilised to optimise the factors. Two stages of optimisation were done using response surface methodology to understand the interaction between factors involved in improving RR120 decolourisation by consortium JR3. Significant factors were screened using Plackett-Burman and further optimisation was done using a central composite design.
2.12.1. Statistical Optimisation Using Plackett-Burman and Central Composite Design
The Plackett-Burman design was chosen to define the most appropriate parameters among the different factors. Design Expert v 13.0 was used for this experiment, along with four parameters selected for a total of 12 experimental runs. Each parameter was analysed at two different levels (low and high) as shown in
Table 3. After a variance analysis was conducted, factors with
p < 0.05 were considered significant and were further optimized using a central composite design.
2.12.2. Optimisation of Significant Factors Using a Central Composite Design
A central composite design was used to analyse the optimal RR120 decolourisation by consortium JR3 and the interaction among the factors with 30 experimental runs. The levels of four significant factors and their levels are illustrated in
Table 4. Based on the conformation run resulting from central composite design (CCD) analysis, an experiment was done to assess the best condition for RR120 decolourisation by consortium JR3.
2.13. Statistical Analysis
All the experiments were conducted in triplicate. Error bars are used to show experimental errors (standard deviation of three determinations). Data from the experiments were statistically analysed using GraphPad v3.5 (GraphPad Software Inc, San Diego, CA, USA) and one-way analysis of variance was done using Tukey’s test.
4. Discussion
A dye obtains its colour through chromophores and auxochromes. By altering the overall electron energy system, the dye loses its colour, hence decolourisation occurs [
4,
32]. However, the term decolourisation is often confused with degradation among researchers. Decolourisation only breaks down the electron system, meanwhile, degradation breaks down the electron system and the resulting compounds are used as energy or nitrogen source [
37]. This has led to various researchers using different media compositions for isolating dye degrading bacteria. Certain media promotes decolourisation while others inhibit degradation.
This is the first study comparing the role of several media in the isolation method and their influence on RR120 decolourisation. Only consortium JR3 was able to decolourise RR120 maximally without the aid of additional carbon sources and co-substrates as compared to consortia JR1 and JR2. This is the first study to report a combination of
Pseudomonas aeruginosa,
Serratia marcescens and
Enterobacter sp. that were able to degrade and consume RR120 as the sole carbon source without the aid of co-substrate. Even though the observed decolourisation was lower than with the addition of extra carbon sources, the consortium can completely mineralise the dye.
Bacillus lentus B1377 [
38] was able to achieve complete mineralisation of RR120 when supplemented with NB, on other hand,
Pseudomonas gualiconensis [
39] was reported to produced toxic metabolite in similar condition. Most of RR120 decolouriser needs glucose/yeast extract more than 3 g/L to achieve significant decolourisation [
16,
26,
30,
39,
40]. Consortium JR3 was able to do so with the least amount of co-substrate.
The process of acclimatisation is more beneficial as it improved the decolourisation capability of JR3. This can be illustrated in MSM 4 when JR3 outperformed JR2 in decolourisation term. Meanwhile, in MSM 3, supplementing a yeast concentration of 1 g/L significantly improved decolourisation ability of JR3 followed by JR2. However, no noticeable decolourisation was observed in JR2 and JR3 when RR120 was supplemented with NB as compared to JR1. Under this condition, RR120 is rather being absorbed by the consortium than being degraded (data not shown). A similar effect was also observed in MSM 2 and MSM 5. Both consortia grow on readily available carbon source and the dye are not being consumed.
The results are in agreement with Brilon et al. [
34]. When no glucose and yeast extract was given to the culture medium, degradation of naphthalenesulfonic acids was extremely poor in
Pseudomonas sp. A3 [
34]. When yeast extract was introduced at a concentration of 0.05% (
w/
v), degradation is improved. They also illustrated that
Pseudomonas sp. C22 that is unable to grow on naphthalenesulfonic acids, is able to grow when the same concentration of yeast extract is introduced [
34]. Thus, it could be summarised that the presence of co-substrate significantly improves decolourisation of RR120 consortium JR3, and without the addition of co-substrate, decolourisation appears to be slow. Consortium JR3 was chosen as the best consortium to decolourise RR120 as it requires less co-substrate and is able to consume RR120 without the presence of any additional carbon source, which is extremely rare in the field of azo dye bioremediation.
Similarly, when co-substrate is not available, the presence of trace elements improves the decolourisation condition. The availability of these nutrients for microbes responsible for aerobic digestion and substrate toxicity. The most important trace elements in micronutrients mostly involved in aerobic digestion efficiency are cobalt, nickel, molybdenum, zinc and magnesium [
41]. Molybdenum functions as cofactors for various enzymes, nickel is needed for the synthesis of coenzymes, zinc is known for stimulating cell growth, iron acts as an electron acceptor, calcium is needed for membrane permeability and both copper and cobalt for metallic enzyme activator [
42].
The role of yeast extract is considered essential for the regeneration of NADH, which acts as the electron donor for the reduction of azo bonds [
43]. Yeast extract comprised components such as riboflavin, thiamine and pyridoxine, which enhance and improve bacterial growth and azo dye decolourising activity. However, the right amount of yeast extract to be used in dye decolourisation remains debatable among researchers. Some researchers found that increasing yeast extract concentration enhances decolourisation rate while others report the opposite [
12,
44]. Consortium JR3 falls into the latter category. This behaviour might be attributed to the fact that bacterial strains consume yeast extract as readily available carbon and nitrogen source for their growth instead of targeting the destruction of azo dye bond. The role of the electron donor taken by RR120 is a result of the fortuitous process [
45]. When this occurs, RR120 are not broken down properly, thus the colour loss will be lower.
Eskandari et al. [
12] reported that ammonium dihydrogen phosphate improved Reactive Blue 5 decolourisation better compared to peptone and yeast extract in both cold-adapted and mesophilic consortium. Organic nitrogen metabolism is crucial for the regeneration of NADH, as it plays an important role as an electron donor [
46]. Organic nitrogen sources, such as peptone, yeast and beef extract, have been found effective in improving dye removal rate in a mesophilic microorganism [
30]. However, excess of this nitrogen source will result in the dye not being properly mineralised, as bacteria start consuming available carbon to grow [
37]. This finding is consistent with the result of Fouda et al. [
47], who reported that ammonium sulphate significantly improved decolourisation of Disperse Blue, Disperse Yellow and Reactive Red Syozol compared to rest of the nitrogen sources in
Pseudomonas stutzeri SB13.
Additionally, dye decolourisation does not indicate that resulting metabolites are less toxic and safe compared to that parent compounds [
48]. An assessment on resulting metabolite should be done to determine the safety of remediated dyes. Seed germination percentage, seedling survival and seedling height have been taken as important criteria to assess plant response to specific pollutants [
7,
35,
39]. The presence of azo dye could directly affect the role of chlorophyllase and abscisic acid (ABA), therefore impeding the growth of the plant [
49]. The severity of dye toxic effect depends on metabolite(s) produced during the decolourisation process. Rawat et al. [
4] found that treating Acid Orange 7 decolourised metabolites significantly reduced the overall shoot biomass of
Allium cepa because of poor uptake and transfer of the nutrient. These metabolites were later detected to be aniline, 1-amino-2-naphthol, naphthalene, and phenyl diazene, the consequences of improver azo dye decolourisation by halophilic microbial consortium. On the other hand, Nouren et al. [
50] reported a better improvement in
Zea mays seed germination, shoot and root length when exposed to treated Direct Yellow 4 compared to untreated samples, which result in a 50% inhibition rate. Similarly, Santana et al. [
51] also observed an increase in lettuce and clove seed germination when exposed to a treated sample of Reactive Red 195 compared to the parent compound as a result of complete mineralisation of the azo dye. The finding of Santana et al. [
51] also supported by the work of Roy et al. [
52] who reported significant improvement in chickpea seed germination when exposed to a treated sample of Reactive Yellow. Therefore, it can be summarised that a proper decolourisation of a dye compound will result in total mineralisation, which significantly reduces the toxicity of the dye.
The nature of the role of pH on dye decolourisation is influenced by the location of the strain obtained. Bacteria that are isolated from the acidic environment are able to degrade dye better in pH less than 7 [
53], meanwhile, those from alkaline environment perform exceptionally well in pH more than 7 [
24]. Previous repeated exposure to acidic or alkaline environment allowed the azoreductase enzyme to acclimatise to their appropriate environment pH; thus, having a distinct affinity towards substrate in different pH environments. Since consortium JR3 was isolated from a contaminated site near to a fabric manufacturing factory, frequent exposure to alkaline effluents from washing activity (detergent and soap) allowed the enzyme to acclimatize to an alkaline condition. Similarly,
Halomonas sp. isolated from textile wash water showed an optimal pH of 9–12 for maximal removal of Reactive Red 152 [
54]. Meanwhile,
Enterococcus casseliflavus RDB4 had an optimal pH of 7 and better decolourisation of Reactive Red 195 is observed under alkaline conditions compared to acidic medium [
55].
Pseudomonas aeruginosa strain HF5 was able to decolourise 90.9% and 82.5% RR120 at pH values of 7.5 and 8.5 at 24 h, respectively [
30]. In most of the reactive red decolourisation works occurring under aerobic alkaline conditions, the detected metabolite is 6-hydroxy cyclohexa 2, 4-dienone, which is a very common desulphonation reaction [
38,
56].
Temperature influences bacterial growth rate and enzymatic reaction. In this study, the optimum temperature was 35 °C. In other studies, the optimum temperature for RR120 removal was 35 °C in
Shewanella haliotis RDB1 [
37]. No decolourisation was observed beyond 55 °C in our result, similar to
Shewanella haliotis RDB1. Consortium RV2 was able to decolourise 87.05% of 100 ppm Reactive Red 31 at the optimal temperature of 37 °C within 12 h under microaerophilic conditions [
57].
The ability of
Pseudomonas sp. SUK1 to decolourise RR120 has been reported [
58]. The strain was able to achieve 97% decolourisation of 150 ppm RR120 at 100 h. However, since nutrient broth and static conditions were used for
Pseudomonas sp. SUK1 decolourisation conditions, resulting in toxic metabolites needing to undergo radiation treatment to breakdown those by-products. Meanwhile,
Pseudomonas guariconensis was able to decolourise a maximum of 48% RR120 at 70 h [
39]. Even in fungal species, decolourisation of RR120 was reported at 100 ppm with the aid of glucose [
59], whereby consortium JR3 was able to decolourise even better without the need for glucose and still achieved significant decolourisation at 400 ppm.
In terms of decolourisation efficiency, RSM resulted in a better decolourisation capability compared to the one-factor-at-a-timeresult. This study also shows similar improvement observed in other degradation optimisation works [
60,
61]. Although both OFAT and RSM showed almost similar values, minor changes in RSM value have significantly improved the decolourisation rate of RR120.