Microbial Profile of the Leachate from Mexico City’s Bordo Poniente Composting Plant: An Inoculum to Digest Organic Waste
Abstract
:1. Introduction
2. Materials and Methods
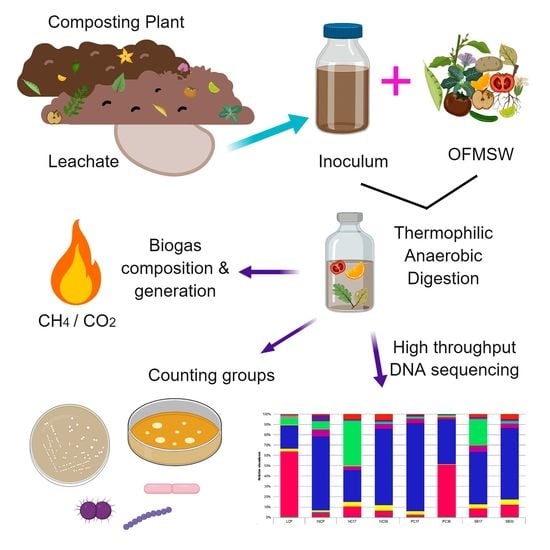
2.1. Sampling and Preparation of Substrate and Inoculum
2.2. Biochemical Methane Potential (BMP) Assay
2.3. Chemical Analysis
2.4. Generation and Composition of Biogas
2.5. Counting Mesophilic and Thermophilic Groups
2.6. Biochemical Tests
2.7. DNA Extraction and 16S rDNA Library Preparation
2.8. High Throughput Sequencing and Analysis
2.9. Bacterial and Archaeal Relative Abundance and Diversity
2.10. Predicted Metabolic Pathways of Bacteria and Archaea
2.11. Statistical Analysis
3. Results
3.1. Chemical Characteristics of the OFMSW, Inoculum, and Reactor Mixtures at the End of the 30-Day Period
3.2. Biochemical Methane Potential Assay
3.3. Counting Mesophilic and Thermophilic Groups
3.4. Bacterial Identification
3.5. Microbial Analysis
3.6. Diversity of Microbial Communities
3.7. Predicted Metabolic Pathways of Bacteria and Archaea
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Martin-Ryals, A.; Schideman, L.; Li, P.; Wilkinson, H.; Wagner, R. Improving anaerobic digestion of a cellulosic waste via routine bioaugmentation with cellulolytic microorganisms. Bioresour. Technol. 2015, 189, 62–70. [Google Scholar] [CrossRef] [PubMed]
- Hoornweg, D.; Bhada, P. What a Waste. A Global Review of Solid Waste Management; World Bank Group: Washington, DC, USA, 2012; p. 44. [Google Scholar]
- Hettiarachchi, H.; Ryu, S.; Caucci, S.; Silva, R. Municipal Solid Waste Management in Latin America and the Caribbean: Issues and Potential Solutions from the Governance Perspective. Recycling 2018, 3, 19. [Google Scholar] [CrossRef]
- Hettiarachchi, H.; Meegoda, J.N.; Ryu, S. Organic Waste Buyback as a Viable Method to Enhance Sustainable Municipal Solid Waste Management in Developing Countries. Int. J. Environ. Res. Public Health 2018, 15, 2483. [Google Scholar] [CrossRef]
- Roubík, H.; Mazancová, J.; Le Dinh, P.; Dinh Van, D.; Banout, J. Biogas quality across small-scale biogas plants: A case of Central Vietnam. Energies 2018, 11, 1794. [Google Scholar] [CrossRef]
- Ghesla, P.L.; Gomes, L.P.; Caetano, M.O.; Miranda, L.A.S.; Dai-Prá, L.B. Municipal solid waste management from the experience of São Leopoldo/Brazil and Zurich/Switzerland. Sustain. 2018, 10. [Google Scholar] [CrossRef]
- Gaceta Oficial de la Ciudad de México. Programa de Gestión Integral de los Residuos Sólidos para la Ciudad de México 2016–2020. 2016, 3–62. Available online: https://data.consejeria.cdmx.gob.mx/portal_old/uploads/gacetas/e2fbcdaadd9665d726ee6bd2195eb913.pdf (accessed on 17 May 2018).
- Garcia-Peña, E.I.; Parameswaran, P.; Kang, D.W.; Canul-Chan, M.; Krajmalnik-Brown, R. Anaerobic digestion and co-digestion processes of vegetable and fruit residues : Process and microbial ecology. Bioresour. Technol. 2011, 102, 9447–9455. [Google Scholar] [CrossRef] [PubMed]
- Lv, W.; Schanbacher, F.L.; Yu, Z. Putting microbes to work in sequence: Recent advances in temperature-phased anaerobic digestion processes. Bioresour. Technol. 2010, 101, 9409–9414. [Google Scholar] [CrossRef] [PubMed]
- Phun, C.; Bong, C.; Yee, L.; Tin, C. The characterisation and treatment of food waste for improvement of biogas production during anaerobic digestion - A review. J. Clean. Prod. 2018, 172, 1545–1558. [Google Scholar] [CrossRef]
- Ariunbaatar, J.; Ozcan, O.; Bair, R.; Esposito, G.; Ball, R.; Lens, P.N.L.; Yeh, D.H. Bioaugmentation of the anaerobic digestion of food waste by dungs of herbivore, carnivore, and omnivore zoo animals. Environ. Technol. 2017, 39, 516–526. [Google Scholar] [CrossRef]
- Lim, J.W.; Chen, C.L.; Ho, I.J.R.; Wang, J.Y. Study of microbial community and biodegradation efficiency for single- and two-phase anaerobic co-digestion of brown water and food waste. Bioresour. Technol. 2013, 147, 193–201. [Google Scholar] [CrossRef]
- Wei, S. The application of biotechnology on the enhancing of biogas production from lignocellulosic waste. Appl. Microbiol. Biotechnol. 2016, 100, 9821–9836. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Wang, H.; Qiu, Y.; Ren, L.; Jiang, B. Microbial characteristics in anaerobic digestion process of food waste for methane production–A review. Bioresour. Technol. 2018, 248, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Venkiteshwaran, K.; Bocher, B.; Maki, J.; Zitomer, D. Relating Anaerobic Digestion Microbial Community and Process Function. Microbiol. Insights 2015, 8, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Boon, N. Inoculum selection is crucial to ensure operational stability in anaerobic digestion. Appl. Microbiol. Biotechnol. 2014, 99, 189–199. [Google Scholar] [CrossRef]
- Koch, K.; Lippert, T.; Drewes, J.E. The role of inoculum’s origin on the methane yield of different substrates in biochemical methane potential (BMP) tests. Bioresour. Technol. 2017, 243, 457–463. [Google Scholar] [CrossRef] [PubMed]
- Dhamodharan, K.; Kumar, V.; Kalamdhad, A.S. Effect of different livestock dungs as inoculum on food waste anaerobic digestion and its kinetics. Bioresour. Technol. 2015, 180, 237–241. [Google Scholar] [CrossRef]
- Mo, Y.; Min, H.; Lee, K.; Chantrasakdakul, P.; Kim, D.; Young, K. Changes in bacterial and archaeal communities in anaerobic digesters treating different organic wastes. Chemosphere 2015, 141, 134–137. [Google Scholar] [CrossRef]
- Yu, Z.; Li, Y. Comparison of different liquid anaerobic digestion effluents as inocula and nitrogen sources for solid state anaerobic digestion of corn stover. Waste Manag. 2015, 33, 26–32. [Google Scholar] [CrossRef]
- Zamanzadeh, M.; Hagen, L.H.; Svensson, K.; Linjordet, R.; Horn, S.J. Anaerobic digestion of food waste - Effect of recirculation and temperature on performance and microbiology. Water Res. 2016, 96, 246–254. [Google Scholar] [CrossRef]
- Zhou, S.; Nikolausz, M.; Zhang, J.; Riya, S.; Terada, A.; Hosomi, M. Variation of the microbial community in thermophilic anaerobic digestion of pig manure mixed with different ratios of rice straw. J. Biosci. Bioeng. 2016, 122, 334–340. [Google Scholar] [CrossRef]
- Ghanimeh, S.; El-Fadel, M.; Saikaly, P. Improving the stability of thermophilic anaerobic digesters treating SS-OFMSW through enrichment with compost and leachate seeds. Bioresour. Technol. 2013, 131, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Guven, H.; Akca, M.S.; Iren, E.; Keles, F.; Ozturk, I.; Altinbas, M. Co-digestion performance of organic fraction of municipal solid waste with leachate: Preliminary studies. Waste Manag. 2018, 71, 775–784. [Google Scholar] [CrossRef] [PubMed]
- Eslami, H.; Hashemi, H.; Fallahzadeh, R.A.; Khosravi, R.; Fard, R.F.; Ebrahimi, A.A. Effect of organic loading rates on biogas production and anaerobic biodegradation of composting leachate in the anaerobic series bioreactors. Ecol. Eng. 2018, 110, 165–171. [Google Scholar] [CrossRef]
- Ming, L.; Xuya, P.; Youcai, Z.; Wenchuan, D.; Huashuai, C.; Guotao, L.; Zhengsong, W. Microbial inoculum with leachate recirculated cultivation for the enhancement of OFMSW composting. J. Hazard. Mater. 2008, 153, 885–891. [Google Scholar] [CrossRef] [PubMed]
- Ramos, P.L.; Kondo, M.Y.; Santos, S.M.B.; de Vasconcellos, S.P.; Rocha, R.C.S.; da Cruz, J.B.; Eugenio, P.F.M.; Cabral, H.; Juliano, M.A.; Juliano, L.; et al. A Tropical Composting Operation Unit at São Paulo Zoo as a Source of Bacterial Proteolytic Enzymes. Appl. Biochem. Biotechnol. 2019, 187, 282–297. [Google Scholar] [CrossRef] [PubMed]
- PAOT. Diagnóstico actual del flujo de residuos sólidos urbanos que se genera en el Distrito Federal 2011, 123. Available online: http://centro.paot.org.mx/documentos/paot/estudios/flujo_residuos_DF.pdf (accessed on 29 May 2019).
- Li, Y.; Nelson, M.C.; Chen, P. Comparison of the microbial communities in solid-state anaerobic digestion ( SS-AD ) reactors operated at mesophilic and thermophilic temperatures. Appl. Microbiol. Biotechnol. 2015, 99, 969–980. [Google Scholar] [CrossRef]
- Shi, J.; Wang, Z.; Stiverson, J.A.; Yu, Z.; Li, Y. Reactor performance and microbial community dynamics during solid-state anaerobic digestion of corn stover at mesophilic and thermophilic conditions. Bioresour. Technol. 2013, 136, 574–581. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Lv, C.; Tong, J.; Liu, J.; Liu, J.; Yu, D.; Wang, Y.; Chen, M.; Wei, Y. Optimization and microbial community analysis of anaerobic co-digestion of food waste and sewage sludge based on microwave pretreatment. Bioresour. Technol. 2016, 200, 253–261. [Google Scholar] [CrossRef]
- Angelidaki, I.; Alves, M.; Bolzonella, D.; Borzacconi, L.; Campos, J.L.; Guwy, A.J.; Kalyuzhnyi, S.; Jenicek, P.; Van Lier, J.B. Defining the biomethane potential (BMP) of solid organic wastes and energy crops: A proposed protocol for batch assays. Water Sci. Technol. 2009, 59, 927–934. [Google Scholar] [CrossRef]
- Sadzawka, A.; Carrasco, M.A.; Grez, R.; Mora, M.d.l.L. Métodos de análisis de compost; Instituto de Investigaciones Agropecuarias, INIA: Santiago, Chile, 2015. [Google Scholar]
- Tale, V.P.; Maki, J.S.; Zitomer, D.H. Bioaugmentation of overloaded anaerobic digesters restores function and archaeal community. Water Res. 2015, 70, 138–147. [Google Scholar] [CrossRef] [Green Version]
- Williams, C.M.; Shih, J.C.H. Enumeration of some microbial groups in thermophilic poultry waste digesters and enrichment of a feather-degrading culture. J. Appl. Bacteriol. 1989, 67, 25–35. [Google Scholar] [CrossRef]
- Murugesan, S.; Ulloa-Martínez, M.; Martínez-Rojano, H.; Galván-Rodríguez, F.M.; Miranda-Brito, C.; Romano, M.C.; Piña-Escobedo, A.; Pizano-Zárate, M.L.; Hoyo-Vadillo, C.; García-Mena, J. Study of the diversity and short-chain fatty acids production by the bacterial community in overweight and obese Mexican children. Eur. J. Clin. Microbiol. Infect. Dis. 2015, 34, 1337–1346. [Google Scholar] [CrossRef] [PubMed]
- Maspolim, Y.; Zhou, Y.; Guo, C.; Xiao, K.; Jern, W. Comparison of single-stage and two-phase anaerobic sludge digestion systems – Performance and microbial community dynamics. Chemosphere 2015, 140, 54–62. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.G.; Han, G.; Lim, J.; Lee, C.; Hwang, S. A comprehensive microbial insight into two-stage anaerobic digestion of food waste-recycling wastewater. Water Res. 2010, 44, 4838–4849. [Google Scholar] [CrossRef] [PubMed]
- Murugesan, S.; Reyes-Mata, M.P.; Nirmalkar, K.; Chavez-Carbajal, A.; Juárez-Hernández, J.I.; Torres-Gómez, R.E.; Piña-Escobedo, A.; Maya, O.; Hoyo-Vadillo, C.; Ramos-Ramírez, E.G.; et al. Profiling of bacterial and fungal communities of Mexican cheeses by high throughput DNA sequencing. Food Res. Int. 2018, 113, 371–381. [Google Scholar] [CrossRef] [PubMed]
- García-Mena, J.; Murugesan, S.; Pérez-Muñoz, A.A.; García-Espitia, M.; Maya, O.; Jacinto-Montiel, M.; Monsalvo-Ponce, G.; Piña-Escobedo, A.; Domínguez-Malfavón, L.; Gómez-Ramírez, M.; et al. Airborne Bacterial Diversity from the Low Atmosphere of Greater Mexico City. Microb. Ecol. 2016, 72, 70–84. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high- throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS One 2013, 8, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Chávez-Carbajal, A.; Nirmalkar, K.; Pérez-Lizaur, A.; Hernández-Quiroz, F.; Ramírez-del-Alto, S.; García-Mena, J.; Hernández-Guerrero, C. Gut Microbiota and Predicted Metabolic Pathways in a Sample of Mexican Women Affected by Obesity and Obesity Plus Metabolic Syndrome. Int. J. Mol. Sci. 2019, 20, 438. [Google Scholar] [CrossRef]
- Tyagi, V.K.; Fdez-Güelfo, L.A.; Zhou, Y.; Álvarez-Gallego, C.J.; Garcia, L.I.R.; Ng, W.J. Anaerobic co-digestion of organic fraction of municipal solid waste (OFMSW): Progress and challenges. Renew. Sustain. Energy Rev. 2018, 93, 380–399. [Google Scholar] [CrossRef]
- Campuzano, R.; Gonzalez-Martinez, S. Characteristics of the organic fraction of municipal solid waste and methane production: A review. Waste Manag. 2016, 54, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Campuzano, R.; González-Martínez, S. Extraction of soluble substances from organic solid municipal waste to increase methane production. Bioresour. Technol. 2015, 178, 247–253. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Li, Y.; Ge, X.; Yang, L.; Li, Y. Anaerobic digestion of food waste – Challenges and opportunities. Bioresour. Technol. 2018, 247, 1047–1058. [Google Scholar] [CrossRef] [PubMed]
- Labatut, R.A.; Angenent, L.T.; Scott, N.R. Biochemical methane potential and biodegradability of complex organic substrates. Bioresour. Technol. 2011, 102, 2255–2264. [Google Scholar] [CrossRef] [PubMed]
- Moset, V.; Poulsen, M.; Højberg, O.; Møller, H.B. Mesophilic versus thermophilic anaerobic digestion of cattle manure : methane productivity and microbial ecology. Microb. Biotechnol. 2015, 8, 787–800. [Google Scholar] [CrossRef] [PubMed]
- Riya, S.; Suzuki, K.; Meng, L.; Zhou, S.; Terada, A.; Hosomi, M. The influence of the total solid content on the stability of dry-thermophilic anaerobic digestion of rice straw and pig manure. Waste Manag. 2018, 76, 350–356. [Google Scholar] [CrossRef]
- Forster-Carneiro, T.; Pérez, M.; Romero, L.I.; Sales, D. Dry-thermophilic anaerobic digestion of organic fraction of the municipal solid waste: Focusing on the inoculum sources. Bioresour. Technol. 2007, 98, 3195–3203. [Google Scholar] [CrossRef]
- Suksong, W.; Mamimin, C.; Prasertsan, P.; Kongjan, P.; O-Thong, S. Effect of inoculum types and microbial community on thermophilic and mesophilic solid-state anaerobic digestion of empty fruit bunches for biogas production. Ind. Crops Prod. 2019, 133, 193–202. [Google Scholar] [CrossRef]
- Raposo, F.; Fernández-Cegrí, V.; de la Rubia, M.A.; Borja, R.; Béline, F.; Cavinato, C.; Demirer, G.; Fernández, B.; Fernández-Polanco, M.; Frigon, J.C.; et al. Biochemical methane potential (BMP) of solid organic substrates: Evaluation of anaerobic biodegradability using data from an international interlaboratory study. J. Chem. Technol. Biotechnol. 2011, 86, 1088–1098. [Google Scholar] [CrossRef]
- Li, W.; Khalid, H.; Zhu, Z.; Zhang, R.; Liu, G.; Chen, C.; Thorin, E. Methane production through anaerobic digestion: Participation and digestion characteristics of cellulose, hemicellulose and lignin. Appl. Energy 2018, 226, 1219–1228. [Google Scholar] [CrossRef]
- Martín-González, L.; Castro, R.; Pereira, M.A.; Alves, M.M.; Font, X.; Vicent, T. Thermophilic co-digestion of organic fraction of municipal solid wastes with FOG wastes from a sewage treatment plant : Reactor performance and microbial community monitoring. Bioresour. Technol. 2011, 102, 4734–4741. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Treu, L.; Kougias, P.G.; Campanaro, S.; Angelidaki, I. Converting mesophilic upflow sludge blanket (UASB) reactors to thermophilic by applying axenic methanogenic culture bioaugmentation. Chem. Eng. J. 2018, 332, 508–516. [Google Scholar] [CrossRef] [Green Version]
- Strang, O.; Ács, N.; Wirth, R.; Maróti, G.; Bagi, Z.; Rákhely, G.; Kovács, K.L. Bioaugmentation of the thermophilic anaerobic biodegradation of cellulose and corn stover. Anaerobe 2017, 46, 104–113. [Google Scholar] [CrossRef] [PubMed]
- Weiss, A.; Jérôme, V.; Freitag, R.; Mayer, H.K. Diversity of the resident microbiota in a thermophilic municipal biogas plant. Appl. Microbiol. Biotechnol. 2008, 81, 163–173. [Google Scholar] [CrossRef] [PubMed]
- Goux, X.; Calusinska, M.; Fossépré, M.; Benizri, E.; Delfosse, P. Start-up phase of an anaerobic full-scale farm reactor - Appearance of mesophilic anaerobic conditions and establishment of the methanogenic microbial community. Bioresour. Technol. 2016, 212, 217–226. [Google Scholar] [CrossRef]
- Sun, W.; Yu, G.; Louie, T.; Liu, T.; Zhu, C.; Xue, G.; Gao, P. From mesophilic to thermophilic digestion: the transitions of anaerobic bacterial, archaeal, and fungal community structures in sludge and manure samples. Appl. Microbiol. Biotechnol. 2015, 99, 10271–10282. [Google Scholar] [CrossRef]
- Ritari, J.; Koskinen, K.; Hultman, J.; Kurola, J.M.; Kymäläinen, M.; Romantschuk, M.; Paulin, L.; Auvinen, P. Molecular analysis of meso- and thermophilic microbiota associated with anaerobic biowaste degradation. BMC Microbiol. 2012, 12, 1–14. [Google Scholar] [CrossRef]
- Heeg, K.; Pohl, M.; Sontag, M.; Mumme, J.; Klocke, M.; Nettmann, E. Microbial communities involved in biogas production from wheat straw as the sole substrate within a two-phase solid-state anaerobic digestion. Syst. Appl. Microbiol. 2014, 37, 590–600. [Google Scholar] [CrossRef]
- Guo, X.; Wang, C.; Sun, F.; Zhu, W.; Wu, W. A comparison of microbial characteristics between the thermophilic and mesophilic anaerobic digesters exposed to elevated food waste loadings. Bioresour. Technol. 2014, 152, 420–428. [Google Scholar] [CrossRef]
- Di Maria, F.; Barratta, M.; Bianconi, F.; Placidi, P.; Passeri, D. Solid anaerobic digestion batch with liquid digestate recirculation and wet anaerobic digestion of organic waste: Comparison of system performances and identification of microbial guilds. Waste Manag. 2017, 59, 172–180. [Google Scholar] [CrossRef]
- Tsapekos, P.; Kougias, P.G.; Vasileiou, S.A.; Treu, L.; Campanaro, S.; Lyberatos, G.; Angelidaki, I. Bioaugmentation with hydrolytic microbes to improve the anaerobic biodegradability of lignocellulosic agricultural residues. Bioresour. Technol. 2017, 234, 350–359. [Google Scholar] [CrossRef] [PubMed]
- Amha, Y.M.; Anwar, M.Z.; Brower, A.; Jacobsen, C.S.; Stadler, L.B.; Webster, T.M.; Smith, A.L. Inhibition of anaerobic digestion processes: Applications of molecular tools. Bioresour. Technol. 2018, 247, 999–1014. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.; Khanal, S.K. A little breath of fresh air into an anaerobic system: How microaeration facilitates anaerobic digestion process. Biotechnol. Adv. 2018, 36, 1971–1983. [Google Scholar] [CrossRef] [PubMed]
- Demirel, B.; Scherer, P. The roles of acetotrophic and hydrogenotrophic methanogens during anaerobic conversion of biomass to methane: A review. Rev. Environ. Sci. Biotechnol. 2008, 7, 173–190. [Google Scholar] [CrossRef]
- Venkiteshwaran, K.; Milferstedt, K.; Hamelin, J.; Zitomer, D.H. Anaerobic digester bioaugmentation influences quasi steady state performance and microbial community. Water Res. 2016, 104, 128–136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goux, X.; Calusinska, M.; Lemaigre, S.; Marynowska, M.; Klocke, M.; Udelhoven, T.; Benizri, E.; Delfosse, P. Microbial community dynamics in replicate anaerobic digesters exposed sequentially to increasing organic loading rate, acidosis, and process recovery. Biotechnol. Biofuels 2015, 8, 122. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Wang, X.; Deng, Y.; He, X.; Li, Z.; Li, Q.; Chen, J.; He, M.; Zhang, M.; Hu, G.; et al. Adaption of microbial community during the start- up stage of a thermophilic anaerobic digester treating food waste. Biosci. Biotechnol. Biochem. 2016, 80, 2025–2032. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, W.; Lee, J.; Loh, K.-C.; Dai, Y.; Tong, Y.W. Enhancement of biogas production in anaerobic co-digestion of food waste and waste activated sludge by biological co-pretreatment. Energy 2017, 1–8. [Google Scholar] [CrossRef]
- Brown, K.; Ghoshdastidar, A.J.; Hanmore, J.; Frazee, J.; Tong, A.Z. Membrane bioreactor technology: A novel approach to the treatment of compost leachate. Waste Manag. 2013, 33, 2188–2194. [Google Scholar] [CrossRef]
- Neumann, L.; Scherer, P. Impact of bioaugmentation by compost on the performance and ecology of an anaerobic digester fed with energy crops. Bioresour. Technol. 2011, 102, 2931–2935. [Google Scholar] [CrossRef]








| Parameters | OFMSW | LCP | INCP |
|---|---|---|---|
| pH | 4.55 ± 0.05 | 6.89 ± 0.03 | 8.33 ± 0.01 |
| ORP (mV) | 47.00 ± 1.73 | −284.00 ± 3.61 | −335.67 ± 3.79 |
| EC (mS/cm) | 17.42 ± 0.92 | 33.35 ± 0.05 | 34.77 ± 0.26 |
| TS (g/kg) | 288.57 ± 34.70 | 56.40 ± 2.56 | 47.70 ± 6.17 |
| VS (g/kg) | 232.44 ± 28.14 | 29.75 ± 1.33 | 23.53 ± 3.30 |
| VS/TS ratio (%) | 80.54 ± 0.14 | 52.76 ± 0.22 | 49.29 ± 1.26 |
| Parameters | NC30 | PC30 | SB30 | p Value |
|---|---|---|---|---|
| pH | 8.51 ± 0.12 | 7.98 ± 0.14 | 7.75 ± 0.16 | 0.001 a |
| ORP (mV) | −366.00 ± 11.53 | −379.00 ± 9.00 | −378.67 ± 3.06 | 0.245 b |
| EC (mS/cm) | 32.17 ± 4.04 | 34.16 ± 3.77 | 40.80 ± 1.35 | 0.041 a |
| TS (g/kg) | 45.68 ± 0.76 | 39.57 ± 0.75 | 43.73 ± 7.08 | 0.258 a |
| VS (g/kg) | 13.90 ± 1.70 | 8.22 ± 0.44 | 19.96 ± 1.78 | 0.004 b |
| VS/TS ratio | 30.41 ± 3.23 | 20.78 ± 0.73 | 46.01 ± 3.89 | 0.004 b |
| Mesophilic (CFU/mL) | Thermophilic (MPN/mL) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| LCP | INCP | Treatments | Day 17 | Day 30 | LCP | INCP | Treatments | Day 17 | Day 30 | |
| TAB | 6.4 × 105 | 5.9 × 105 | NC | 3.7 × 104 | 5.3 × 105 | 4.3 × 104 | 4.3 × 104 | NC | 9.5 × 104 | 2.9 × 103 |
| PC | 3.5 × 104 | 3.2 × 105 | PC | 2.4 × 105 | 1.5 × 104 | |||||
| SB | 3.7 × 104 | 2.8 × 105 | SB | 2.4 × 105 | 1.6 × 103 | |||||
| CA | 4.6 × 106 | 8.9 × 104 | NC | 9.1 × 104 | 6.9 × 106 | 35 | 1.1 × 103 | NC | 3.5 × 103 | 3.5 × 103 |
| PC | 4.9 × 107 | 1.5 × 105 | PC | 3.5 × 103 | 2.1 × 104 | |||||
| SB | 6.1 × 106 | 8.2 × 104 | SB | 1.5 × 103 | 2.0 × 103 | |||||
| Sample | Taxa (%) | ||||
|---|---|---|---|---|---|
| LCP | g_Bacteroides (40.29) | f_Porphyromonadaceae (18.59) | g_Acidaminococcus (3.35) | f_Lachnospiraceae (2.48) | Others (35.30) |
| INCP | o_MBA08 (47.07) | c_OPB54 (6.95) | o_ML615J-2 (5.59) | g_Acinetobacter (4.86) | Others (35.52) |
| NC17 | g_Acinetobacter (36.25) | o_MBA08 (10.65) | g_S1 (4.91) | f_Porphyromonadaceae (3.90) | Others (44.28) |
| NC30 | o_MBA08 (46.02) | g_S1 (5.31) | c_OPB54 (4.58) | f_Porphyromonadaceae (4.21) | Others (39.88) |
| PC17 | f_Lachnospiraceae (42.57) | f_Halanaerobiaceae (11.03) | o_MBA08 (8.71) | f_Ruminococcaceae (6.07) | Others (31.63) |
| PC30 | g_Bacteroides (26.82) | f_S24-7 (13.61) | f_Ruminococcaceae (10.21) | f_Lachnospiraceae (5.38) | Others (39.03) |
| SB17 | f_Pseudomonadaceae (18.73) | o_MBA08 (13.60) | o_OPB54 (8.13) | c_OPB54 (6.19) | Others (53.35) |
| SB30 | o_MBA08 (30.29) | o_OPB54 (8.66) | o_Bacteroidales (8.00) | f_Halanaerobiaceae (5.17) | Others (47.88) |
| Sample | Taxa (%) | |||
|---|---|---|---|---|
| LCP | g_Methanosarcina (81.09) | g_Methanobrevibacter (16.10) | g_Methanoculleus (0.63) | Others (2.17) |
| INCP | g_Methanoculleus (77.28) | g_Methanosarcina (15.42) | g_Methanomassiliicoccus (3.33) | Others (7.30) |
| NC17 | g_vadinCA11 (54.08) | g_Methanoculleus (35.87) | g_Methanobrevibacter (3.47) | Others (6.58) |
| NC30 | g_vadinCA11 (65.45) | g_Methanoculleus (25.60) | g_Methanosarcina (2.9681.09) | Others (6.00) |
| PC17 | g_Methanoculleus (92.57) | g_vadinCA11 (4.19) | g_Methanosarcina (1.62) | Others (1.61) |
| PC30 | g_Methanoculleus (75.07) | g_Methanothermobacter (11.35) | g_vadinCA11 (7.22) | Others (6.35) |
| SB17 | g_Methanoculleus (91.67) | g_Methanosarcina (3.71) | g_vadinCA11 (2.44) | Others (2.17) |
| SB30 | g_Methanoculleus (59.79) | g_vadinCA11 (29.24) | g_Methanosarcina (5.55) | Others (5.42) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gállego Bravo, A.K.; Salcedo Serrano, D.A.; López Jiménez, G.; Nirmalkar, K.; Murugesan, S.; García-Mena, J.; Gutiérrez Castillo, M.E.; Tovar Gálvez, L.R. Microbial Profile of the Leachate from Mexico City’s Bordo Poniente Composting Plant: An Inoculum to Digest Organic Waste. Energies 2019, 12, 2343. https://0-doi-org.brum.beds.ac.uk/10.3390/en12122343
Gállego Bravo AK, Salcedo Serrano DA, López Jiménez G, Nirmalkar K, Murugesan S, García-Mena J, Gutiérrez Castillo ME, Tovar Gálvez LR. Microbial Profile of the Leachate from Mexico City’s Bordo Poniente Composting Plant: An Inoculum to Digest Organic Waste. Energies. 2019; 12(12):2343. https://0-doi-org.brum.beds.ac.uk/10.3390/en12122343
Chicago/Turabian StyleGállego Bravo, Aixa Kari, Daniel Alejandro Salcedo Serrano, Gloria López Jiménez, Khemlal Nirmalkar, Selvasankar Murugesan, Jaime García-Mena, María Eugenia Gutiérrez Castillo, and Luis Raúl Tovar Gálvez. 2019. "Microbial Profile of the Leachate from Mexico City’s Bordo Poniente Composting Plant: An Inoculum to Digest Organic Waste" Energies 12, no. 12: 2343. https://0-doi-org.brum.beds.ac.uk/10.3390/en12122343