Bioengineered Fluorescent Nanoprobe Conjugates for Tracking Human Bone Cells: In Vitro Biocompatibility Analysis
Abstract
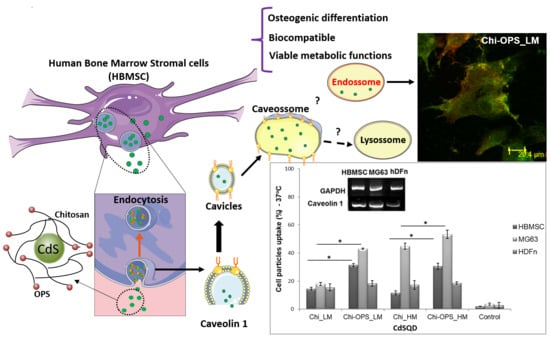
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Bioconjugation of the Peptides (OPS) to Chitosan (Chi)
2.3. Production of CdS/Polysaccharide Conjugates
2.4. Chemic-Physical Evaluation of CdS/Polysaccharide Conjugates
2.5. Biological Characterization of CdS Conjugates
2.5.1. Cell Viability Assay
2.5.2. Flow Cytometry Single-Cell Analysis (FACS)
2.5.3. Vesicular Trafficking Evaluation
2.5.4. Osteogenic Differentiation Evaluation
2.5.5. Osteogenic Differentiation-Related Gene Expresion (RT-PCR)
2.5.6. Different mRNA Expression of Caveolin-1 (Real-Time PCR)
2.5.7. Hemolytic Activity
2.6. Statistical Analysis
3. Results
3.1. In Vitro Evaluation of CdS/Polysaccharide Conjugates
3.2. Fluorescent Bio-Nanoprobes Biocompatibility Evaluation
3.3. Cellular Uptake of QD Nanoconjugates
3.4. HBMSC’s Osteogenic Differentiation Evaluation
3.5. Hemocompatibility Assay
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- White, E.S.; Mantovani, A.R. Inflammation, wound repair, and fibrosis: Reassessing the spectrum of tissue injury and resolution. J. Pathol. 2013, 229, 141–144. [Google Scholar] [CrossRef] [PubMed]
- Chan, B.P.; Leong, K.W. Scaffolding in tissue engineering: General approaches and tissue-specific considerations. Eur. Spine J. 2008, 17 (Suppl. 4), 467–479. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.; Cui, Z.K.; Fan, J.; Fartash, A.; Aghaloo, T.L.; Lee, M. Photocrosslinkable chitosan hydrogels functionalized with the RGD peptide and phosphoserine to enhance osteogenesis. J. Mater. Chem. B 2016, 4, 5289–5298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, P.; Zheng, J.; Zhang, Z.; Hou, Y.; Wang, J.; Zhang, C.; Quan, C. Bioactive 3D scaffolds self-assembled from phosphorylated mimicking peptide amphiphiles to enhance osteogenesis. J. Biomater. Sci. Polym. Ed. 2018, 30, 34–48. [Google Scholar] [CrossRef]
- Raucci, M.G.; Alvarez-Perez, M.A.; Meikle, S.; Ambrosio, L.; Santin, M. Poly (Epsilon-lysine) dendrons tethered with phosphoserine increase mesenchymal stem cell differentiation potential of calcium phosphate gels. Tissue Eng. Part A 2014, 20, 474–485. [Google Scholar] [CrossRef] [PubMed]
- Zilberman, Y.; Kallai, I.; Gafni, Y.; Pelled, G.; Kossodo, S.; Yared, W.; Gazit, D. Fluorescence molecular tomography enables in vivo visualization and quantification of nonunion fracture repair induced by genetically engineered mesenchymal stem cells. J. Orthop. Res. 2008, 26, 522–530. [Google Scholar] [CrossRef] [PubMed]
- Mansur, H.S.; Gonzalez, J.C.; Mansur, A.A. Biomolecule-quantum dot systems for bioconjugation applications. Colloids Surf. B Biointerfaces 2011, 84, 360–368. [Google Scholar] [CrossRef]
- Mansur, H.S. Quantum dots and nanocomposites. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2010, 2, 113–129. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Pang, J.; Xu, F.; Zhang, X. Simple Approach to Synthesize Amino-Functionalized Carbon Dots by Carbonization of Chitosan. Sci. Rep. 2016, 6, 31100. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Liu, H.; Wang, L.; Su, X. A label-free fluorescence biosensor for highly sensitive detection of lectin based on carboxymethyl chitosan-quantum dots and gold nanoparticles. Anal. Chim. Acta 2016, 932, 88–97. [Google Scholar] [CrossRef]
- Chen, S.; Chen, X.; Xia, T.; Ma, Q. A novel electrochemiluminescence sensor for the detection of nitroaniline based on the nitrogen-doped graphene quantum dots. Biosens. Bioelectron. 2016, 85, 903–908. [Google Scholar] [CrossRef] [PubMed]
- Mansur, H.S.; Mansur, A.A.P.; Curti, E.; De Almeida, M.V. Functionalized-chitosan/quantum dot nano-hybrids for nanomedicine applications: Towards biolabeling and biosorbing phosphate metabolites. J. Mater. Chem. B 2013, 1, 1696–1711. [Google Scholar] [CrossRef]
- Mansur, H.S.; Piscitelli Mansur, A.A. Fluorescent nanohybrids: Quantum dots coupled to polymer recombinant protein conjugates for the recognition of biological hazards. J. Mater. Chem. 2012, 22, 9006–9018. [Google Scholar] [CrossRef]
- Hossain, M.A.; Chowdhury, T.; Bagul, A. Imaging modalities for the in vivo surveillance of mesenchymal stromal cells. J. Tissue Eng. Regen. Med. 2015, 9, 1217–1224. [Google Scholar] [CrossRef]
- Ruan, Y.; Yu, W.; Cheng, F.; Zhang, X.; Larre, S. Detection of prostate stem cell antigen expression in human prostate cancer using quantum-dot-based technology. Sensors 2012, 12, 5461–5470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oh, E.; Liu, R.; Nel, A.; Gemill, K.B.; Bilal, M.; Cohen, Y.; Medintz, I.L. Meta-analysis of cellular toxicity for cadmium-containing quantum dots. Nat. Nanotechnol. 2016, 11, 479–486. [Google Scholar] [CrossRef]
- Tsoi, K.M.; Dai, Q.; Alman, B.A.; Chan, W.C. Are quantum dots toxic? Exploring the discrepancy between cell culture and animal studies. Acc. Chem. Res. 2013, 46, 662–671. [Google Scholar] [CrossRef]
- Olsen, J.V.; Blagoev, B.; Gnad, F.; Macek, B.; Kumar, C.; Mortensen, P.; Mann, M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 2006, 127, 635–648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salgado, C.L.; Teixeira, B.I.B.; Monteiro, F.J.M. Biomimetic Composite Scaffold with Phosphoserine Signaling for Bone Tissue Engineering Application. Front. Front. Bioeng. Biotechnol. 2019, 7, 206. [Google Scholar] [CrossRef]
- Salgado, C.L.; Barrias, C.C.; Monteiro, F.J.M. Clarifying the Tooth-Derived Stem Cells Behavior in a 3D Biomimetic Scaffold for Bone Tissue Engineering Applications. Front. Bioeng. Biotechnol. 2020, 8, 724. [Google Scholar] [CrossRef]
- Bailey, T.J.; Fossum, S.L.; Fimbel, S.M.; Montgomery, J.E.; Hyde, D.R. The inhibitor of phagocytosis, O-phospho-L-serine, suppresses Muller glia proliferation and cone cell regeneration in the light-damaged zebrafish retina. Exp. Eye Res. 2010, 91, 601–612. [Google Scholar] [CrossRef] [Green Version]
- Gaitonde, P.; Purohit, V.S.; Balu-Iyer, S.V. Intravenous administration of Factor VIII-O-Phospho-L-Serine (OPLS) complex reduces immunogenicity and preserves pharmacokinetics of the therapeutic protein. Eur. J. Pharm. Sci. 2015, 66, 157–162. [Google Scholar] [CrossRef] [Green Version]
- Salgado, C.L.; Mansur, A.A.P.; Mansur, H.S.; Monteiro, F.J.M. Fluorescent bionanoprobes based on quantum dot-chitosan-O-phospho-l-serine conjugates for labeling human bone marrow stromal cells. RSC Adv. 2014, 4, 49016–49027. [Google Scholar] [CrossRef]
- Rajeshwar, K.; de Tacconi, N.R.; Chenthamarakshan, C.R. Semiconductor-Based Composite Materials: Preparation, Properties, and Performance. Chem. Mater. 2001, 13, 2765–2782. [Google Scholar] [CrossRef]
- Tauc, J.; Menth, A. States in the gap. J. Non-Cryst. Solids 1972, 8, 569–585. [Google Scholar] [CrossRef]
- Spanhel, L.; Haase, M.; Weller, H.; Henglein, A. Photochemistry of colloidal semiconductors. 20. Surface modification and stability of strong luminescing CdS particles. J. Am. Chem. Soc. 1987, 109, 5649–5655. [Google Scholar] [CrossRef]
- Hocaoglu, I.; Asik, D.; Ulusoy, G.; Grandfils, C.; Ojea-Jimenez, I.; Rossi, F.; Kiraz, A.; Doğan, N.; Acar, H.Y. Cyto/hemocompatible magnetic hybrid nanoparticles (Ag2S-Fe3O4) with luminescence in the near-infrared region as promising theranostic materials. Colloids Surf. B Biointerfaces 2015, 133, 198–207. [Google Scholar] [CrossRef]
- Mansur, A.A.; Mansur, H.S.; de Carvalho, S.M.; Lobato, Z.I.; Guedes, M.I.; Leite, M.F. Surface biofunctionalized CdS and ZnS quantum dot nanoconjugates for nanomedicine and oncology: To be or not to be nanotoxic? Int. J. Nanomed. 2016, 11, 4669–4690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rejman, J.; Oberle, V.; Zuhorn, I.S.; Hoekstra, D. Size-dependent internalization of particles via the pathways of clathrin- and caveolae-mediated endocytosis. Biochem. J. 2004, 377, 159–169. [Google Scholar] [CrossRef] [PubMed]
- Elkin, S.R.; Lakoduk, A.M.; Schmid, S.L. Endocytic pathways and endosomal trafficking: A primer. Wiener Medizinische Wochenschrift (1946) 2016, 166, 196–204. [Google Scholar] [CrossRef] [Green Version]
- Rzigalinski, B.A.; Strobl, J.S. Cadmium-containing nanoparticles: Perspectives on pharmacology and toxicology of quantum dots. Toxicol. Appl. Pharmacol. 2009, 238, 280–288. [Google Scholar] [CrossRef] [Green Version]
- Baker, N.; Tuan, R.S. The less-often-traveled surface of stem cells: Caveolin-1 and caveolae in stem cells, tissue repair and regeneration. Stem Cell Res. Ther. 2013, 4, 90. [Google Scholar] [CrossRef] [Green Version]
- Baker, N.; Zhang, G.; You, Y.; Tuan, R.S. Caveolin-1 regulates proliferation and osteoinductivedifferentiation of human mesenchymal stem cells. J. Cell. Biochem. 2012, 113, 3773–3787. [Google Scholar] [CrossRef]
- Fielding, C.J.; Fielding, P.E. Cholesterol and caveolae: Structural and functional relationships. Biochim. Et Biophys. Acta 2000, 1529, 210–222. [Google Scholar] [CrossRef]
- Koleske, A.J.; Baltimore, D.; Lisanti, M.P. Reduction of caveolin and caveolae in oncogenically transformed cells. Proc. Natl. Acad. Sci. USA 1995, 92, 1381–1385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.W.; Reimer, C.L.; Oh, P.; Campbell, D.B.; Schnitzer, J.E. Tumor cell growth inhibition by caveolin re-expression in human breast cancer cells. Oncogene 1998, 16, 1391–1397. [Google Scholar] [CrossRef] [Green Version]
- Abbah, S.A.; Lu, W.W.; Chan, D.; Cheung, K.M.C.; Liu, W.G.; Zhao, F.; Li, Z.Y.; Leong, J.C.Y.; Luk, K.D.K. Osteogenic behavior of alginate encapsulated bone marrow stromal cells: An in vitro study. J. Mater. Sci. Mater. Med. 2008, 19, 2113–2119. [Google Scholar] [CrossRef]
- Healy, K.E.; Guldberg, R.E. Bone tissue engineering. J. Musculoskelet. Neuronal Interact. 2007, 7, 328–330. [Google Scholar] [PubMed]
- Moioli, E.K.; Hong, L.; Mao, J.J. Inhibition of osteogenic differentiation of human mesenchymal stem cells. Wound Repair Regen. 2007, 15, 413–421. [Google Scholar] [CrossRef] [PubMed]
- Stein, G.S.; Lian, J.B.; Owen, T.A. Relationship of cell growth to the regulation of tissue-specific gene expression during osteoblast differentiation. FASEB J. 1990, 4, 3111–3123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neve, A.; Corrado, A.; Cantatore, F.P. Osteoblast physiology in normal and pathological conditions. Cell Tissue Res. 2011, 343, 289–302. [Google Scholar] [CrossRef] [PubMed]
- Chenu, C.; Colucci, S.; Grano, M.; Zigrino, P.; Barattolo, R.; Zambonin, G.; Baldini, N.; Vergnaud, P.; Delmas, P.D.; Zallone, A.Z. Osteocalcin induces chemotaxis, secretion of matrix proteins, and calcium-mediated intracellular signaling in human osteoclast-like cells. J. Cell Biol. 1994, 127, 1149–1158. [Google Scholar] [CrossRef] [PubMed]
- Lacey, D.L.; Timms, E.; Tan, H.L.; Kelley, M.J.; Dunstan, C.R.; Burgess, T.; Elliott, R.; Colombero, A.; Elliott, G.; Scully, S.; et al. Osteoprotegerin ligand is a cytokine that regulates osteoclast differentiation and activation. Cell. 1998, 93, 165–176. [Google Scholar] [CrossRef] [Green Version]
- De Carvalho, S.M.; Mansur, A.A.P.; Mansur, H.S.; Guedes, M.I.M.C.; Lobato, Z.I.P.; Leite, M.F. In vitro and in vivo assessment of nanotoxicity of CdS quantum dot/aminopolysaccharide bionanoconjugates. Mater. Sci. Eng. C 2017, 71, 412–424. [Google Scholar] [CrossRef] [PubMed]













| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| GAPDH | 5’-TAACTGGTAAAGTGGATATTG-3’ | 5’-GAAGATGGTAGATGGATTTC-3’ |
| Runx-2 | 5’-GTGCCTAGGCGCATTTCA-3’ | 5’-GCTCTTCTTACTGAGATGGAAGG-3’ |
| OC | 5’-AGAGTCCAGCAAAGGTGCAG-3’ | 5’-TCAGCCAACTCGTCACAGTC-3’ |
| ALP | 5’-ACGTGGCTAAGAATGTCATC-3’ | 5’-CTGGTAGGCGATGTCCTTA-3’ |
| BMP-2 | 5’-GACGAGGTCCTGAGCGAGTT-3’ | 5’-GCAATGGCCTTATCTGTGAC-3’ |
| OPN | 5′-ACTCGAACGACTCTGATGATGT-3′ | 5′-GTCAGGTCTGCGAAACTTCTTA-3′ |
| OPG | 5’-AAGGAGCTGCAGTACGTCAA-3’ | 5’-CTGCTCGAAGGTGAGGTTAG-3’ |
| Caveol1 | 5’-AACAACCCGAACATCTACAACGGG-3′ | 5’-AAGGACTAACTCTAAGTCACGTAGTC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salgado, C.L.; Mansur, A.A.P.; Mansur, H.S.; Monteiro, F.J.M. Bioengineered Fluorescent Nanoprobe Conjugates for Tracking Human Bone Cells: In Vitro Biocompatibility Analysis. Materials 2021, 14, 4422. https://0-doi-org.brum.beds.ac.uk/10.3390/ma14164422
Salgado CL, Mansur AAP, Mansur HS, Monteiro FJM. Bioengineered Fluorescent Nanoprobe Conjugates for Tracking Human Bone Cells: In Vitro Biocompatibility Analysis. Materials. 2021; 14(16):4422. https://0-doi-org.brum.beds.ac.uk/10.3390/ma14164422
Chicago/Turabian StyleSalgado, Christiane L., Alexandra A. P. Mansur, Herman S. Mansur, and Fernando J. M. Monteiro. 2021. "Bioengineered Fluorescent Nanoprobe Conjugates for Tracking Human Bone Cells: In Vitro Biocompatibility Analysis" Materials 14, no. 16: 4422. https://0-doi-org.brum.beds.ac.uk/10.3390/ma14164422