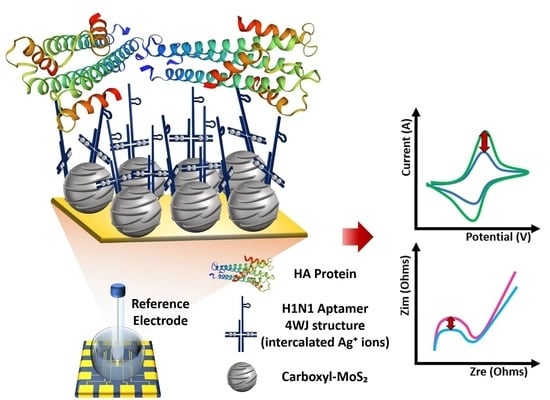
Fabrication of Electrochemical Influenza Virus (H1N1) Biosensor Composed of Multifunctional DNA Four-Way Junction and Molybdenum Disulfide Hybrid Material
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Synthesis of Carboxyl-MoS2
2.3. Assembly of Multifunctional DNA 4WJ
2.4. Fabrication of Electrode
2.5. Fabrication of Multifunctional DNA 4WJ/Carboxyl-MoS2 Heterolayer
2.6. Surface Morphology Analysis
2.7. Electrochemical Analysis of Multifunctional 4WJ/Carboxyl-MoS2 Heterolayer on Electrode
3. Results
3.1. Construction of Multifunctional DNA 4WJ
3.2. Investigation of Fabricated HA Protein/Multifunctional DNA 4WJ on Carboxyl-MoS2 Heterolayer
3.3. Electrochemical Response of H1N1 Detection
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Neumann, G.; Noda, T.; Kawaoka, Y. Emergence and pandemic potential of swine-origin H1N1 influenza virus. Nature 2009, 459, 931–939. [Google Scholar] [CrossRef] [Green Version]
- Iuliano, A.D.; Roguski, K.M.; Chang, H.H.; Muscatello, D.J.; Palekar, R.; Tempia, S.; Cohen, C.; Gran, J.M.; Schanzer, D.; Cowling, B.J.; et al. Estimates of global seasonal influenza-associated respiratory mortality: A modelling study. Lancet 2018, 391, 1285–1300. [Google Scholar] [CrossRef]
- Imai, M.; Watanabe, T.; Hatta, M.; Das, S.C.; Ozawa, M.; Shinya, K.; Zhong, G.; Hanson, A.; Katsura, H.; Watanabe, S.; et al. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature 2012, 486, 420–428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yeh, E.; Luo, R.F.; Dyner, L.; Hong, D.K.; Banaei, N.; Baron, E.J.; Pinsky, B.A. Preferential lower respiratory tract infection in swine-origin 2009 A(HlNl) influenza. Clin. Infect. Dis. 2010, 50, 391–394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Girard, M.P.; Tam, J.S.; Assossou, O.M.; Kieny, M.P. The 2009 A (H1N1) influenza virus pandemic: A review. Vaccine 2010, 28, 4895–4902. [Google Scholar] [PubMed]
- Calitri, C.; Gabiano, C.; Garazzino, S.; Pinon, M.; Zoppo, M.; Cuozzo, M.; Scolfaro, C.; Tovo, P.A. Clinical features of hospitalised children with 2009 H1N1 influenza virus infection. Eur. J. Pediatr. 2010, 169, 1511–1515. [Google Scholar] [CrossRef] [PubMed]
- Writing Committee of the WHO Consultation on Clinical Aspects of Pandemic (H1N1) 2009 Influenza. Clinical aspects of pandemic 2009 influenza A (H1N1) virus infection. N. Engl. J. Med. 2010, 362, 1708–1719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vemula, S.V.; Zhao, J.; Liu, J.; Wang, X.; Biswas, S.; Hewlett, I. Current approaches for diagnosis of influenza virus infections in humans. Viruses 2016, 8, 96. [Google Scholar] [CrossRef] [Green Version]
- Ginocchio, C.C.; George, K.S. Likelihood that an unsubtypeable influenza A virus result obtained with the luminex xTAG respiratory virus panel is indicative of infection with novel A/H1N1 (swine-like) influenza virus. J. Clin. Microbiol. 2009, 47, 2347–2348. [Google Scholar] [CrossRef] [Green Version]
- Ganzenmueller, T.; Kluba, J.; Hilfrich, B.; Puppe, W.; Verhagen, W.; Heim, A.; Schulz, T.; Henke-Gendo, C. Comparison of the performance of direct fluorescent antibody staining, a point-of-care rapid antigen test and virus isolation with that of RT-PCR for the detection of novel 2009 influenza A (H1N1) virus in respiratory specimens. J. Med. Microbiol. 2010, 59, 713–717. [Google Scholar] [CrossRef] [Green Version]
- Fouchier, R.A.; Schneeberger, P.M.; Rozendaal, F.W.; Broekman, J.M.; Kemink, S.A.; Munster, V.; Kuiken, T.; Rimmelzwaan, G.F.; Schutten, M.; Van Doornum, G.J.; et al. Avian influenza A virus (H7N7) associated with human conjunctivitis and a fatal case of acute respiratory distress syndrome. Proc. Natl. Acad. Sci. USA 2004, 101, 1356–1361. [Google Scholar] [PubMed] [Green Version]
- Blyth, C.C.; Iredell, J.R.; Dwyer, D.E. Rapid-Test Sensitivity for Novel Swine-Origin Influenza A (H1N1) Virus in Humans. N. Engl. J. Med. 2009, 361, 2493. [Google Scholar] [CrossRef] [PubMed]
- Ginocchio, C.C.; Zhang, F.; Manji, R.; Arora, S.; Bornfreund, M.; Falk, L.; Lotlikar, M.; Kowerska, M.; Becker, G.; Korologos, D.; et al. Evaluation of multiple test methods for the detection of the novel 2009 influenza A (H1N1) during the New York City outbreak. J. Clin. Virol. 2009, 45, 191–195. [Google Scholar] [CrossRef] [PubMed]
- Vasoo, S.; Stevens, J.; Singh, K. Rapid antigen tests for diagnosis of pandemic (Swine) influenza A/H1N1. Clin. Infect. Dis. 2009, 49, 1090–1093. [Google Scholar] [CrossRef]
- Krejcova, L.; Hynek, D.; Michalek, P.; Milosavljevic, V.; Kopel, P.; Zitka, O.; Konecna, M.; Kynicky, J.; Adam, V.; Hubalek, J.; et al. Electrochemical sensors and biosensors for influenza detection—Literature survey 2012–2013. Int. J. Electrochem. Sci. 2014, 9, 3440–3448. [Google Scholar]
- Tepeli, Y.; Ülkü, A. Electrochemical biosensors for influenza virus a detection: The potential of adaptation of these devices to POC systems. Sens. Actuators B Chem. 2018, 254, 377–384. [Google Scholar] [CrossRef]
- Diouani, M.F.; Helali, S.; Hafaid, I.; Hassen, W.M.; Snoussi, M.A.; Ghram, A.; Jaffrezic-Renault, N.; Abdelghani, A. Miniaturized biosensor for avian influenza virus detection. Mater. Sci. Eng. C 2008, 28, 580–583. [Google Scholar] [CrossRef]
- Lilley, D.M.; Clegg, R.M. The structure of the four-way junction in DNA. Ann. Rev. Biophys. Biomol. Struct. 1993, 22, 299–328. [Google Scholar] [CrossRef] [PubMed]
- Clegg, R.M.; Zechel, A.; Carlberg, C.; Diekmann, S.; Murchie, A.I.H.; Lilley, D.M.J. Fluorescence Resonance Energy Transfer Analysis of the Structure of the Four-Way DNA Junction. Biochemistry 1992, 31, 4846–4856. [Google Scholar] [CrossRef]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Xiang, D.; Shigdar, S.; Qiao, G.; Wang, T.; Kouzani, A.Z.; Zhou, S.F.; Kong, L.; Li, Y.; Pu, C.; Duan, W. Nucleic acid aptamer-guided cancer therapeutics and diagnostics: The next generation of cancer medicine. Theranostics 2015, 5, 23–42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhardwaj, J.; Chaudhary, N.; Kim, H.; Jang, J. Subtyping of influenza A H1N1 virus using a label-free electrochemical biosensor based on the DNA aptamer targeting the stem region of HA protein. Anal. Chim. Acta 2019, 1064, 94–103. [Google Scholar] [CrossRef] [PubMed]
- Famulok, M.; Mayer, G. Intramers and aptamers: Applications in protein-function analyses and potential for drug screening. ChemBioChem 2005, 6, 19–26. [Google Scholar] [CrossRef]
- Ono, A.; Cao, S.; Togashi, H.; Tashiro, M.; Fujimoto, T.; MacHinami, T.; Oda, S.; Miyake, Y.; Okamoto, I.; Tanaka, Y. Specific interactions between silver(i) ions and cytosine-cytosine pairs in DNA duplexes. Chem. Commun. 2008, 4825–4827. [Google Scholar] [CrossRef]
- Mohammadniaei, M.; Yoon, J.; Lee, T.; Bharate, B.G.; Jo, J.; Lee, D.; Choi, J.W. Electrochemical Biosensor Composed of Silver Ion-Mediated dsDNA on Au-Encapsulated Bi2Se3 Nanoparticles for the Detection of H2O2 Released from Breast Cancer Cells. Small 2018, 14, 1–13. [Google Scholar] [CrossRef]
- Yoon, J.; Mohammadniaei, M.; Choi, H.K.; Shin, M.; Bapurao, G.B.; Lee, T.; Choi, J.W. Resistive switching biodevice composed of MoS2-DNA heterolayer on the gold electrode. Appl. Surf. Sci. 2019, 478, 134–141. [Google Scholar] [CrossRef]
- Cooper, J.P.; Hagerman, P.J. Gel electrophoretic analysis of the geometry of a DNA four-way junction. J. Mol. Biol. 1987, 198, 711–719. [Google Scholar] [CrossRef]
- Lee, T.; Mohammadniaei, M.; Zhang, H.; Yoon, J.; Choi, H.K.; Guo, S.; Guo, P.; Choi, J.W. Single Functionalized pRNA/Gold Nanoparticle for Ultrasensitive MicroRNA Detection Using Electrochemical Surface-Enhanced Raman Spectroscopy. Adv. Sci. 2020, 7, 1902477. [Google Scholar] [CrossRef]
- Lee, T.; Kim, G.H.; Kim, S.M.; Hong, K.; Kim, Y.; Park, C.; Sohn, H.; Min, J. Label-free localized surface plasmon resonance biosensor composed of multi-functional DNA 3 way junction on hollow Au spike-like nanoparticles (HAuSN) for avian influenza virus detection. Coll. Surf. B Biointerfaces 2019, 182, 110341. [Google Scholar] [CrossRef]
- Park, S.Y.; Kim, J.; Yim, G.; Jang, H.; Lee, Y.; Kim, S.M.; Park, C.; Lee, M.H.; Lee, T. Fabrication of electrochemical biosensor composed of multi-functional DNA/rhodium nanoplate heterolayer for thyroxine detection in clinical sample. Coll. Surf. B Biointerfaces 2020, 195, 111240. [Google Scholar] [CrossRef] [PubMed]
- Kwon, J.; Lee, Y.; Lee, T.; Ahn, J.H. Aptamer-based field-effect transistor for detection of avian influenza virus in chicken serum. Anal. Chem. 2020, 92, 5524–5531. [Google Scholar] [CrossRef] [PubMed]
- Mazrouei, R.; Huffman, B.; Sumita, M.; Shavezipur, M. Development of an impedance-based interdigitated biochemical sensor using a multiuser silicon process. J. Micromech. Microeng. 2019, 29. [Google Scholar] [CrossRef]
- Chang, Y.F.; Wang, S.F.; Huang, J.C.; Su, L.C.; Yao, L.; Li, Y.C.; Wu, S.C.; Chen, Y.M.A.; Hsieh, J.P.; Chou, C. Detection of swine-origin influenza A (H1N1) viruses using a localized surface plasmon coupled fluorescence fiber-optic biosensor. Biosens. Bioelectron. 2010, 26, 1068–1073. [Google Scholar] [CrossRef]
- Su, L.C.; Chang, C.M.; Tseng, Y.L.; Chang, Y.F.; Li, Y.C.; Chang, Y.S.; Chou, C. Rapid and highly sensitive method for influenza A (H1N1) virus detection. Anal. Chem. 2012, 84, 3914–3920. [Google Scholar] [CrossRef]
- Zhang, J.; Tian, J.; He, Y.; Chen, S.; Jiang, Y.; Zhao, Y.; Zhao, S. Protein-binding aptamer assisted signal amplification for the detection of influenza A (H1N1) DNA sequences based on quantum dot fluorescence polarization analysis. Analyst 2013, 138, 4722–4727. [Google Scholar] [CrossRef]
- Lee, N.; Wang, C.; Park, J. User-friendly point-of-care detection of influenza A (H1N1) virus using light guide in three-dimensional photonic crystal. RSC Adv. 2018, 8, 22991–22997. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, S.R.; Kim, J.; Suzuki, T.; Lee, J.; Park, E.Y. Detection of influenza virus using peroxidase-mimic of gold nanoparticles. Biotechnol. Bioeng. 2016, 113, 2298–2303. [Google Scholar] [CrossRef] [Green Version]
- Singh, R.; Hong, S.; Jang, J. Label-free Detection of Influenza Viruses using a Reduced Graphene Oxide-based Electrochemical Immunosensor Integrated with a Microfluidic Platform. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef] [Green Version]






| DNA | DNA Sequence |
|---|---|
| H1N1 HA aptamer-tagged 4WJ-a | 5′-TAC TGC ACA CGA CAC CGA CTG TCA CCA TCA CCT CGG CGC AGT GCT GGA ACT CCC CGA CTG C-3′ |
| Amine group-tagged 4WJ-b | 5′-CCA GTC CCC CAG CCC ACA TAC TTT GTT GAT CC-3′-NH2 |
| 4WJ-c | 5′-GGA TCA ATC ACC CCT GGC AA-3′ |
| 4WJ-d | 5′-TTG CCA CCC CTG TGT ATG TGG GTT CCA GCA C-3′ |
| Probe | Detection Method | LOD | Linear Range | Ref. |
|---|---|---|---|---|
| Antibody | LSP | 13.9 pg/mL | 5 ~ 50 ng/mL | [34] |
| Antibody | SPR | 30 PFU/mL | Not in the linear range | [35] |
| DNA Aptamer | Fluorescence | 3.45 nM | 10 ~ 100 nM | [36] |
| DNA Aptamer | CV/EIS | 3.7 PFU/mL | 10 ~ 10,000 PFU/mL | [23] |
| DNA Aptamer | Fluorescence | 138 pg/mL | 200 pg/mL ~ 200 μg/mL | [37] |
| Antibody/Antigen | Absorbance | 10.79 pg/mL | 10 pg/mL ~ 10 μg/mL | [38] |
| Antibody | CV/EIS | 0.5 PFU/mL | 1 ~ 10,000 PFU/mL | [39] |
| DNA Aptamer | CV/EIS | 10 pM | 10 pM ~ 100 nM | Present Study |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, J.A.; Kim, J.; Kim, S.M.; Sohn, H.; Park, C.; Kim, T.-H.; Lee, J.-H.; Lee, M.-H.; Lee, T. Fabrication of Electrochemical Influenza Virus (H1N1) Biosensor Composed of Multifunctional DNA Four-Way Junction and Molybdenum Disulfide Hybrid Material. Materials 2021, 14, 343. https://0-doi-org.brum.beds.ac.uk/10.3390/ma14020343
Park JA, Kim J, Kim SM, Sohn H, Park C, Kim T-H, Lee J-H, Lee M-H, Lee T. Fabrication of Electrochemical Influenza Virus (H1N1) Biosensor Composed of Multifunctional DNA Four-Way Junction and Molybdenum Disulfide Hybrid Material. Materials. 2021; 14(2):343. https://0-doi-org.brum.beds.ac.uk/10.3390/ma14020343
Chicago/Turabian StylePark, Jeong Ah, Jinmyeong Kim, Soo Min Kim, Hiesang Sohn, Chulhwan Park, Tae-Hyung Kim, Jin-Ho Lee, Min-Ho Lee, and Taek Lee. 2021. "Fabrication of Electrochemical Influenza Virus (H1N1) Biosensor Composed of Multifunctional DNA Four-Way Junction and Molybdenum Disulfide Hybrid Material" Materials 14, no. 2: 343. https://0-doi-org.brum.beds.ac.uk/10.3390/ma14020343