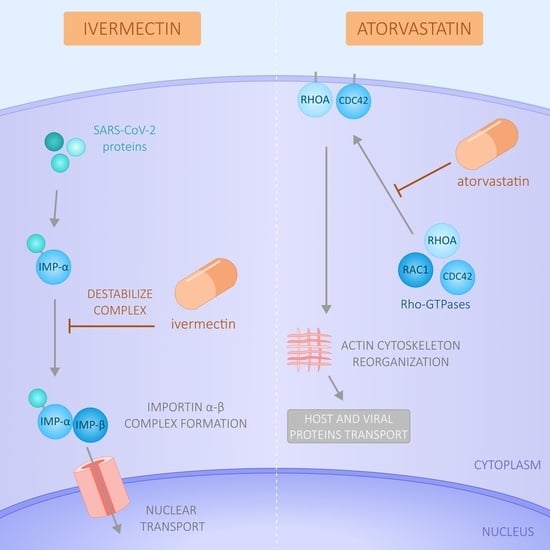
Effect of Ivermectin and Atorvastatin on Nuclear Localization of Importin Alpha and Drug Target Expression Profiling in Host Cells from Nasopharyngeal Swabs of SARS-CoV-2- Positive Patients
Abstract
:1. Introduction
2. Materials and Methods
2.1. Analysis of Publicly Available RNA-Seq Datasets
2.2. Drugs and Reagents
2.3. Cell Lines and Culture Conditions
2.4. Poly(I:C) Treatment
2.5. RNA Isolation, c-DNA Synthesis, and Quantitative Real-Time PCR (RT-qPCR)
2.6. Cell Viability Assay
2.7. Immunofluorescence Staining of Importin α
2.8. Confocal Microscopy Studies and Acquisition Settings
2.9. Statistics
3. Results
3.1. Expression of the Importin Family Genes in SARS-CoV-2-Positive and -Negative Patients
3.2. Expression of the Rho GTPase Family Genes in SARS-CoV-2-Positive and -Negative Patients
3.3. Gene Correlation Analysis in SARS-CoV-2-Positive and -Negative Patients
3.4. Modulation of Importin and Rho GTPases Transcriptional Expression by IVM and ATV in a Viral Infection Simulation Context
3.5. Effect of IVM and ATV on Importin α Nuclear Accumulation
3.6. Combinational Effect of IVM Plus ATV Treatment on Importin α Nuclear Accumulation
3.7. Reduction of Importin α Nuclear Accumulation Using a >10-Fold Lower IVM Concentration
3.8. Reduction of Importin α Nuclear Accumulation Using IVM at Low Concentration in Combination with ATV
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pushpakom, S.; Iorio, F.; Eyers, P.A.; Escott, K.J.; Hopper, S.; Wells, A.; Doig, A.; Guilliams, T.; Latimer, J.; McNamee, C.; et al. Drug repurposing: Progress, challenges and recommendations. Nat. Rev. Drug Discov. 2019, 18, 41–58. [Google Scholar] [CrossRef] [PubMed]
- Serafin, M.B.; Bottega, A.; Foletto, V.S.; da Rosa, T.F.; Hörner, A.; Hörner, R. Drug repositioning is an alternative for the treatment of coronavirus COVID-19. Int. J. Antimicrob. Agents 2020, 55, 105969. [Google Scholar] [CrossRef]
- Alonso, D.F.; Farina, H.G. Repurposing of host-based therapeutic agents for the treatment of coronavirus disease 2019 (COVID-19): A link between antiviral and anticancer mechanisms? Int. J. Antimicrob. Agents 2020, 56, 106125. [Google Scholar] [CrossRef] [PubMed]
- Jans, D.A.; Wagstaff, K.M. Ivermectin as a Broad-Spectrum Host-Directed Antiviral: The Real Deal? Cells 2020, 9, 2100. [Google Scholar] [CrossRef]
- Wagstaff, K.M.; Sivakumaran, H.; Heaton, S.M.; Harrich, D.; Jans, D.A. Ivermectin is a specific inhibitor of importin α/β-mediated nuclear import able to inhibit replication of HIV-1 and dengue virus. Biochem. J. 2012, 443, 851–856. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caly, L.; Druce, J.D.; Catton, M.G.; Jans, D.A.; Wagstaff, K.M. The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antivir. Res. 2020, 178, 104787. [Google Scholar] [CrossRef]
- Krolewiecki, A.; Lifschitz, A.; Moragas, M.; Travacio, M.; Valentini, R.; Alonso, D.F.; Solari, R.; Tinelli, M.A.; Cimino, R.O.; Álvarez, L.; et al. Antiviral effect of high-dose ivermectin in adults with COVID-19: A proof-of-concept randomized trial. EClinicalMedicine 2021, 37, 100959. [Google Scholar] [CrossRef]
- Farina, H.G.; Bublik, D.R.; Alonso, D.F.; Gomez, D.E. Lovastatin alters cytoskeleton organization and inhibits experimental metastasis of mammary carcinoma cells. Clin. Exp. Metastasis 2002, 19, 551–559. [Google Scholar] [CrossRef]
- Chi, X.; Wang, S.; Huang, Y.; Stamnes, M.; Chen, J.-L. Roles of Rho GTPases in Intracellular Transport and Cellular Transformation. Int. J. Mol. Sci. 2013, 14, 7089–7108. [Google Scholar] [CrossRef]
- Bizzotto, J.; Sanchis, P.; Abbate, M.; Lage-Vickers, S.; Lavignolle, R.; Toro, A.; Olszevicki, S.; Sabater, A.; Cascardo, F.; Vazquez, E.; et al. SARS-CoV-2 Infection Boosts MX1 Antiviral Effector in COVID-19 Patients. iScience 2020, 23, 101585. [Google Scholar] [CrossRef]
- Lieberman, N.A.P.; Peddu, V.; Xie, H.; Shrestha, L.; Huang, M.L.; Mears, M.C.; Cajimat, M.N.; Bente, D.A.; Shi, P.Y.; Bovier, F.; et al. In vivo antiviral host transcriptional response to SARS-CoV-2 by viral load, sex, and age. PLoS Biol. 2020, 18, e3000849. [Google Scholar] [CrossRef]
- Blanco-Melo, D.; Nilsson-Payant, B.E.; Liu, W.C.; Uhl, S.; Hoagland, D.; Møller, R.; Jordan, T.X.; Oishi, K.; Panis, M.; Sachs, D.; et al. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell 2020, 181, 1036–1045.e1039. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- World Health Organization. Novel Coronavirus (2019-nCoV) Situation Report-1. 2020. Available online: https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200121-sitrep-1-2019-ncov.pdf?sfvrsn=20a99c10_4 (accessed on 15 June 2021).
- Sanchis, P.; Lavignolle, R.; Abbate, M.; Lage-Vickers, S.; Vazquez, E.; Cotignola, J.; Bizzotto, J.; Gueron, G. Analysis workflow of publicly available RNA-sequencing datasets. STAR Protoc. 2021, 2, 100478. [Google Scholar] [CrossRef]
- Boerma, M.; Fu, Q.; Wang, J.; Loose, D.S.; Bartolozzi, A.; Ellis, J.L.; McGonigle, S.; Paradise, E.; Sweetnam, P.; Fink, L.M.; et al. Comparative gene expression profiling in three primary human cell lines after treatment with a novel inhibitor of Rho kinase or atorvastatin. Blood Coagul. Fibrinolysis Int. J. Haemost. Thromb. 2008, 19, 709–718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Garona, J.; Pifano, M.; Pastrian, M.B.; Gomez, D.E.; Ripoll, G.V.; Alonso, D.F. Addition of vasopressin synthetic analogue [V(4)Q(5)]dDAVP to standard chemotherapy enhances tumour growth inhibition and impairs metastatic spread in aggressive breast tumour models. Clin. Exp. Metastasis 2016, 33, 589–600. [Google Scholar] [CrossRef] [Green Version]
- Garona, J.; Pifano, M.; Orlando, U.D.; Pastrian, M.B.; Iannucci, N.B.; Ortega, H.H.; Podesta, E.J.; Gomez, D.E.; Ripoll, G.V.; Alonso, D.F. The novel desmopressin analogue [V4Q5]dDAVP inhibits angiogenesis, tumour growth and metastases in vasopressin type 2 receptor-expressing breast cancer models. Int. J. Oncol. 2015, 46, 2335–2345. [Google Scholar] [CrossRef] [PubMed]
- Poon, I.K.; Oro, C.; Dias, M.M.; Zhang, J.; Jans, D.A. Apoptin nuclear accumulation is modulated by a CRM1-recognized nuclear export signal that is active in normal but not in tumor cells. Cancer Res. 2005, 65, 7059–7064. [Google Scholar] [CrossRef] [Green Version]
- Tay, M.Y.; Fraser, J.E.; Chan, W.K.; Moreland, N.J.; Rathore, A.P.; Wang, C.; Vasudevan, S.G.; Jans, D.A. Nuclear localization of dengue virus (DENV) 1-4 non-structural protein 5; protection against all 4 DENV serotypes by the inhibitor Ivermectin. Antivir. Res. 2013, 99, 301–306. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Iglesias, Á.; Fuente, R.; Gil-Peña, H.; Alonso-Duran, L.; García-Bengoa, M.; Santos, F.; López, J.M. A simple method based on confocal microscopy and thick sections recognizes seven subphases in growth plate chondrocytes. Sci. Rep. 2020, 10, 6935. [Google Scholar] [CrossRef] [PubMed]
- Audsley, M.D.; Jans, D.A.; Moseley, G.W. Nucleocytoplasmic trafficking of Nipah virus W protein involves multiple discrete interactions with the nuclear import and export machinery. Biochem. Biophys. Res. Commun. 2016, 479, 429–433. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, S.C.; Audsley, M.D.; Lieu, K.G.; Marsh, G.A.; Thomas, D.R.; Heaton, S.M.; Paxman, J.J.; Wagstaff, K.M.; Buckle, A.M.; Moseley, G.W.; et al. Recognition by host nuclear transport proteins drives disorder-to-order transition in Hendra virus V. Sci. Rep. 2018, 8, 358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wickham, H. ggplot2. Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Kassambara, A. “Ggplot2” Based Publication Ready Plots; R Package Version 0.4.0.; Available online: https://CRAN.R-project.org/package=ggpubr” (accessed on 15 June 2021).
- Schloerke, B.; Cook, D.; Larmarange, J.; Francois, B.; Marbach, M.; Thoen, E.; Elberg, A.; Crowley, J. GGally: Extension to “ggplot2”. R Packag Version 2.1.2.; Available online: https://CRAN.R-project.org/package=GGally. (accessed on 15 June 2021).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019; Available online: http://www.R-project.org/ (accessed on 15 June 2021).
- Huang, L.Y.; Stuart, C.; Takeda, K.; D’Agnillo, F.; Golding, B. Poly(I:C) Induces Human Lung Endothelial Barrier Dysfunction by Disrupting Tight Junction Expression of Claudin-5. PLoS ONE 2016, 11, e0160875. [Google Scholar] [CrossRef]
- Zeidler, A.; Karpinski, T.M. SARS-CoV, MERS-CoV, SARS-CoV-2 Comparison of Three Emerging Coronaviruses. Jundishapur J. Microbiol. 2020, 13, e103744. [Google Scholar] [CrossRef]
- Miyamoto, Y.; Saiwaki, T.; Yamashita, J.; Yasuda, Y.; Kotera, I.; Shibata, S.; Shigeta, M.; Hiraoka, Y.; Haraguchi, T.; Yoneda, Y. Cellular stresses induce the nuclear accumulation of importin alpha and cause a conventional nuclear import block. J. Cell Biol. 2004, 165, 617–623. [Google Scholar] [CrossRef] [Green Version]
- Menna, P.L.; Parera, R.L.; Cardama, G.A.; Alonso, D.F.; Gomez, D.E.; Farina, H.G. Enhanced cytostatic activity of statins in mouse mammary carcinoma cells overexpressing β2-chimaerin. Mol. Med. Rep. 2009, 2, 97–102. [Google Scholar] [CrossRef] [Green Version]
- Bello, M. Elucidation of the inhibitory activity of ivermectin with host nuclear importin α and several SARS-CoV-2 targets. J. Biomol. Struct. Dyn. 2021, 1–9. [Google Scholar] [CrossRef]
- Shukla, D.A.K.; Misra, D.S. Antiviral Effects of Ivermectin in COVID-19- Clinically Plausible? Int. J. Infect. Dis. IJID Off. Publ. Int. Soc. Infect. Dis. 2021, 109, 91. [Google Scholar] [CrossRef]
- Siedner, M.J. Ivermectin for the treatment of COVID-19 disease: Too good to pass up or too good to be true? Open Forum Infect. Dis. 2021, ofab318. [Google Scholar] [CrossRef]
- Guzzo, C.A.; Furtek, C.I.; Porras, A.G.; Chen, C.; Tipping, R.; Clineschmidt, C.M.; Sciberras, D.G.; Hsieh, J.Y.; Lasseter, K.C. Safety, tolerability, and pharmacokinetics of escalating high doses of ivermectin in healthy adult subjects. J. Clin. Pharmacol. 2002, 42, 1122–1133. [Google Scholar] [CrossRef]
- Kern, C.; Schöning, V.; Chaccour, C.; Hammann, F. Modeling of SARS-CoV-2 Treatment Effects for Informed Drug Repurposing. Front. Pharmacol. 2021, 12, 625678. [Google Scholar] [CrossRef]
- López-Medina, E.; López, P.; Hurtado, I.C.; Dávalos, D.M.; Ramirez, O.; Martínez, E.; Díazgranados, J.A.; Oñate, J.M.; Chavarriaga, H.; Herrera, S.; et al. Effect of Ivermectin on Time to Resolution of Symptoms Among Adults With Mild COVID-19: A Randomized Clinical Trial. JAMA 2021, 325, 1426–1435. [Google Scholar] [CrossRef] [PubMed]
- Errecalde, J.; Lifschitz, A.; Vecchioli, G.; Ceballos, L.; Errecalde, F.; Ballent, M.; Marín, G.; Daniele, M.; Turic, E.; Spitzer, E.; et al. Safety and Pharmacokinetic Assessments of a Novel Ivermectin Nasal Spray Formulation in a Pig Model. J. Pharm. Sci. 2021, 110, 2501–2507. [Google Scholar] [CrossRef] [PubMed]
- Gower, T.L.; Peeples, M.E.; Collins, P.L.; Graham, B.S. RhoA Is Activated During Respiratory Syncytial Virus Infection. Virology 2001, 283, 188–196. [Google Scholar] [CrossRef] [PubMed]
- Lever, A.R.; Park, H.; Mulhern, T.J.; Jackson, G.R.; Comolli, J.C.; Borenstein, J.T.; Hayden, P.J.; Prantil-Baun, R. Comprehensive evaluation of poly(I:C) induced inflammatory response in an airway epithelial model. Physiol. Rep. 2015, 3, e12334. [Google Scholar] [CrossRef] [PubMed]
- Harris, P.; Sridhar, S.; Peng, R.; Phillips, J.E.; Cohn, R.G.; Burns, L.; Woods, J.; Ramanujam, M.; Loubeau, M.; Tyagi, G.; et al. Double-stranded RNA induces molecular and inflammatory signatures that are directly relevant to COPD. Mucosal Immunol. 2013, 6, 474–484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fortier, M.E.; Kent, S.; Ashdown, H.; Poole, S.; Boksa, P.; Luheshi, G.N. The viral mimic, polyinosinic:polycytidylic acid, induces fever in rats via an interleukin-1-dependent mechanism. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2004, 287, R759–R766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, T.; Zhao, S.; Jin, G.; Song, M.; Zhi, Y.; Zhao, R.; Ma, F.; Zheng, Y.; Wang, K.; Liu, H.; et al. Cytokine Signature Induced by SARS-CoV-2 Spike Protein in a Mouse Model. Front. Immunol. 2020, 11, 621441. [Google Scholar] [CrossRef]
- Hill, A.; Garratt, A.; Levi, J.; Falconer, J.; Ellis, L.; McCann, K.; Pilkington, V.; Qavi, A.; Wang, J.; Wentzel, H. Expression of Concern: “Meta-analysis of Randomized Trials of Ivermectin to Treat SARS-CoV-2 Infection”. Open Forum Infect. Dis. 2021, 8, ofab394. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Song, Y.; Ci, X.; An, N.; Ju, Y.; Li, H.; Wang, X.; Han, C.; Cui, J.; Deng, X. Ivermectin inhibits LPS-induced production of inflammatory cytokines and improves LPS-induced survival in mice. Inflamm. Res. 2008, 57, 524–529. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.; Ci, X.; Chen, N.; Chen, C.; Li, X.; Chu, X.; Li, J.; Deng, X. Anti-inflammatory effects of ivermectin in mouse model of allergic asthma. Inflamm. Res. 2011, 60, 589–596. [Google Scholar] [CrossRef] [PubMed]
- de Melo, G.D.; Lazarini, F.; Larrous, F.; Feige, L.; Kornobis, E.; Levallois, S.; Marchio, A.; Kergoat, L.; Hardy, D.; Cokelaer, T.; et al. Attenuation of clinical and immunological outcomes during SARS-CoV-2 infection by ivermectin. EMBO Mol. Med. 2021, 13, e14122. [Google Scholar] [CrossRef]
- Fedson, D.S.; Opal, S.M.; Rordam, O.M. Hiding in Plain Sight: An Approach to Treating Patients with Severe COVID-19 Infection. mBio 2020, 11, e00398-20. [Google Scholar] [CrossRef] [Green Version]
- Haji Aghajani, M.; Moradi, O.; Azhdari Tehrani, H.; Amini, H.; Pourheidar, E.; Hatami, F.; Rabiei, M.M.; Sistanizad, M. Promising effects of atorvastatin on mortality and need for mechanical ventilation in patients with severe COVID-19; a retrospective cohort study. Int. J. Clin. Pract. 2021, 75, e14434. [Google Scholar] [CrossRef]







| Gene | Forward (5′–3′) | Reverse (5′–3′) | T° An. |
|---|---|---|---|
| PPIA | GGTATAAAAGGGGCGGGAGG | CTGCAAACAGCTCAAAGGAGAC | 60° |
| NFKB | ACTCGCCACCCGGCTTCAGA | GGGCCATCTGCTGTTGGCAGT | 60° |
| KPNA7 | AGGACATGGAGCTGAGAAGTC | GACTGACCGCCATCCTCTG | 57° |
| KPNA5 | GCATTAAGGGCAGTTGGT | CAGTCCAGCAGGCTTCTTT | 57° |
| KPNA2 | GTGGACCCTTTGAACGCAGT | TTGAATCTGTGAAGACGGGCA | 60° |
| RHOA | AAGGACCAGTTCCCAGAGGT | AGCCAACTCTACCTGCTTTCC | 58° |
| RAC1 | CCCCCTATCCTATCCGCAAAC | AACACATCGGCAATCGGCTT | 58° |
| CDC42 | AGGCTGTCAAGTATGTGGAGTG | TCTTCTTCGGTTCTGGAGGC | 60° |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Segatori, V.I.; Garona, J.; Caligiuri, L.G.; Bizzotto, J.; Lavignolle, R.; Toro, A.; Sanchis, P.; Spitzer, E.; Krolewiecki, A.; Gueron, G.; et al. Effect of Ivermectin and Atorvastatin on Nuclear Localization of Importin Alpha and Drug Target Expression Profiling in Host Cells from Nasopharyngeal Swabs of SARS-CoV-2- Positive Patients. Viruses 2021, 13, 2084. https://0-doi-org.brum.beds.ac.uk/10.3390/v13102084
Segatori VI, Garona J, Caligiuri LG, Bizzotto J, Lavignolle R, Toro A, Sanchis P, Spitzer E, Krolewiecki A, Gueron G, et al. Effect of Ivermectin and Atorvastatin on Nuclear Localization of Importin Alpha and Drug Target Expression Profiling in Host Cells from Nasopharyngeal Swabs of SARS-CoV-2- Positive Patients. Viruses. 2021; 13(10):2084. https://0-doi-org.brum.beds.ac.uk/10.3390/v13102084
Chicago/Turabian StyleSegatori, Valeria Inés, Juan Garona, Lorena Grisel Caligiuri, Juan Bizzotto, Rosario Lavignolle, Ayelén Toro, Pablo Sanchis, Eduardo Spitzer, Alejandro Krolewiecki, Geraldine Gueron, and et al. 2021. "Effect of Ivermectin and Atorvastatin on Nuclear Localization of Importin Alpha and Drug Target Expression Profiling in Host Cells from Nasopharyngeal Swabs of SARS-CoV-2- Positive Patients" Viruses 13, no. 10: 2084. https://0-doi-org.brum.beds.ac.uk/10.3390/v13102084