A Hepatitis B Virus-Derived Peptide Can Inhibit Infection of Human Lung Cells with SARS-CoV-2 in a Type-1 Interferon-Dependent Manner
Abstract
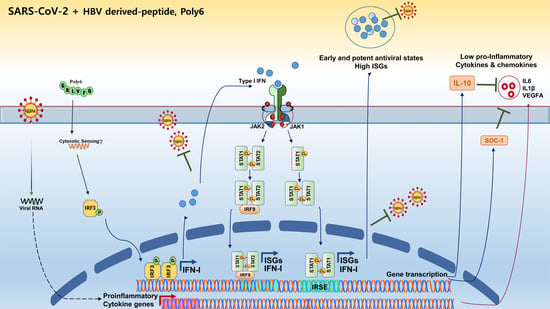
:1. Introduction
2. Materials and Methods
2.1. Viruses and Cell Cultures
2.2. Compounds and Reagents
2.3. Infections
2.4. Quantification of SARS-CoV-2 Viral RNA Genome Copy Number by RT-qPCR
2.5. Cell Cytotoxicity Assay
2.6. Immunofluorescence and Dose–Response Curve (DRC) Analysis
2.7. IFN-I Neutralization Assay
2.8. Western Blot Analysis
2.9. Antibodies
2.10. Flow Cytometric Analysis of Nucleocapsid Protein Expression
2.11. Time-of-Addition Assay
2.12. qPCR Array Analysis
2.13. Enzyme-Linked Immunosorbent Assay (ELISA)
2.14. Plaque Reduction Assay in Vero-E6 Cells
2.15. Quantification and Statistical Analysis
3. Results
3.1. Poly6 Inhibits SARS-CoV-2 Infection in Calu-3 but Not in the Vero-E6 Cell Line in a Dose-Dependent Manner
3.2. Immunofluorescence and Dose–Response Curve Analysis Showed a Potent Inhibitory Effect of Poly6 against Nucleocapsid Protein (NP)
3.3. Poly6 Displayed Limited Ability to Control SARS-CoV-2 in Vero E6 Cells
3.4. Poly6 Exerts an Anti-SARS-CoV-2 Effect in an IFN-I-Dependent Manner in Calu-3 Cells
3.5. Poly6 Inhibits Pro-Inflammatory Cytokine Production in SARS-CoV-2-Infected Calu-3 Cells in an IFN-I-Dependent Manner
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cui, J.; Li, F.; Shi, Z.-L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019, 17, 181–192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, B.; Guo, H.; Zhou, P.; Shi, Z.L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Blanco-Melo, D.; Nilsson-Payant, B.E.; Liu, W.C.; Uhl, S.; Hoagland, D.; Moller, R.; Jordan, T.X.; Oishi, K.; Panis, M.; Sachs, D.; et al. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell 2020, 181, 1036–1045.e9. [Google Scholar] [CrossRef]
- Lazear, H.M.; Schoggins, J.W.; Diamond, M.S. Shared and Distinct Functions of Type I and Type III Interferons. Immunity 2019, 50, 907–923. [Google Scholar] [CrossRef] [PubMed]
- Jakimovski, D.; Kolb, C.; Ramanathan, M.; Zivadinov, R.; Weinstock-Guttman, B. Interferon β for multiple sclerosis. Cold Spring Harb. Perspect. Med. 2018, 8, a032003. [Google Scholar] [CrossRef]
- Wang, N.; Zhan, Y.; Zhu, L.; Hou, Z.; Liu, F.; Song, P.; Qiu, F.; Wang, X.; Zou, X.; Wan, D. Retrospective multicenter cohort study shows early interferon therapy is associated with favorable clinical responses in COVID-19 patients. Cell Host Microbe 2020, 28, 455–464.e2. [Google Scholar] [CrossRef]
- Ribero, M.S.; Jouvenet, N.; Dreux, M.; Nisole, S. Interplay between SARS-CoV-2 and the type I interferon response. PLoS Pathogens 2020, 16, e1008737. [Google Scholar] [CrossRef]
- Davoudi-Monfared, E.; Rahmani, H.; Khalili, H.; Hajiabdolbaghi, M.; Salehi, M.; Abbasian, L.; Kazemzadeh, H.; Yekaninejad, M.S. A randomized clinical trial of the efficacy and safety of interferon β-1a in treatment of severe COVID-19. Antimicrob. Agents Chemother. 2020, 64, e01061-20. [Google Scholar] [CrossRef]
- Trouillet-Assant, S.; Viel, S.; Gaymard, A.; Pons, S.; Richard, J.C.; Perret, M.; Villard, M.; Brengel-Pesce, K.; Lina, B.; Mezidi, M.; et al. Type I IFN immunoprofiling in COVID-19 patients. J. Allergy Clin. Immunol. 2020, 146, 206–208.e2. [Google Scholar] [CrossRef]
- Simon, D.; Tascilar, K.; Krönke, G.; Kleyer, A.; Zaiss, M.M.; Heppt, F.; Meder, C.; Atreya, R.; Klenske, E.; Dietrich, P. Patients with immune-mediated inflammatory diseases receiving cytokine inhibitors have low prevalence of SARS-CoV-2 seroconversion. Nat. Commun. 2020, 11, 1–7. [Google Scholar] [CrossRef]
- Kim, H.; Lee, S.-A.; Kim, D.-W.; Lee, S.-H.; Kim, B.-J. Naturally occurring mutations in large surface genes related to occult infection of hepatitis B virus genotype C. PLoS ONE 2013, 8, e54486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mun, H.S.; Lee, S.A.; Jee, Y.; Kim, H.; Park, J.H.; Song, B.C.; Yoon, J.H.; Kim, Y.J.; Lee, H.S.; Hyun, J.W. The prevalence of hepatitis B virus preS deletions occurring naturally in Korean patients infected chronically with genotype C. J. Med. Virol. 2008, 80, 1189–1194. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-A.; Kim, K.-J.; Kim, H.; Choi, W.-H.; Won, Y.-S.; Kim, B.-J. Hepatitis B virus preS1 deletion is related to viral replication increase and disease progression. World J. Gastroenterol. 2015, 21, 5039. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Lee, S.Y.; Choi, Y.M.; Kim, B.J. HBV polymerase-derived peptide exerts an anti-HIV-1 effect by inhibiting the acetylation of viral integrase. Biochem. Biophys. Res. Commun. 2018, 501, 541–546. [Google Scholar] [CrossRef]
- Yang, S.-B.; Lee, M.-H.; Kim, B.-R.; Choi, Y.-M.; Kim, B.-J. A Hepatitis B Virus-Derived Peptide Exerts an Anticancer Effect via TNF/iNOS-producing Dendritic Cells in Tumor-Bearing Mouse Model. Cancers 2021, 13, 407. [Google Scholar] [CrossRef]
- Hoffmann, M.; Mösbauer, K.; Hofmann-Winkler, H.; Kaul, A.; Kleine-Weber, H.; Krüger, N.; Gassen, N.C.; Müller, M.A.; Drosten, C.; Pöhlmann, S. Chloroquine does not inhibit infection of human lung cells with SARS-CoV-2. Nature 2020, 585, 588–590. [Google Scholar] [CrossRef]
- Onabajo, O.O.; Banday, A.R.; Stanifer, M.L.; Yan, W.; Obajemu, A.; Santer, D.M.; Florez-Vargas, O.; Piontkivska, H.; Vargas, J.M.; Ring, T.J. Interferons and viruses induce a novel truncated ACE2 isoform and not the full-length SARS-CoV-2 receptor. Nat. Genet. 2020, 52, 1283–1293. [Google Scholar] [CrossRef]
- Tang, Y.; Liu, J.; Zhang, D.; Xu, Z.; Ji, J.; Wen, C. Cytokine storm in COVID-19: The current evidence and treatment strategies. Front. Immunol. 2020, 11, 1708. [Google Scholar] [CrossRef]
- Chen, G.; Wu, D.; Guo, W.; Cao, Y.; Huang, D.; Wang, H.; Wang, T.; Zhang, X.; Chen, H.; Yu, H.; et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Investig. 2020, 130, 2620–2629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [Green Version]
- Giamarellos-Bourboulis, E.J.; Netea, M.G.; Rovina, N.; Akinosoglou, K.; Antoniadou, A.; Antonakos, N.; Damoraki, G.; Gkavogianni, T.; Adami, M.E.; Katsaounou, P.; et al. Complex Immune Dysregulation in COVID-19 Patients with Severe Respiratory Failure. Cell Host Microbe 2020, 27, 992–1000.e3. [Google Scholar] [CrossRef]
- Zhang, Q.; Bastard, P.; Liu, Z.; Le Pen, J.; Moncada-Velez, M.; Chen, J.; Ogishi, M.; Sabli, I.K.; Hodeib, S.; Korol, C. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science 2020, 370, 6515. [Google Scholar] [CrossRef]
- Bastard, P.; Rosen, L.B.; Zhang, Q.; Michailidis, E.; Hoffmann, H.H.; Zhang, Y.; Dorgham, K.; Philippot, Q.; Rosain, J.; Beziat, V.; et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science 2020, 370. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Zhao, Y.; Zhang, F.C.; Wang, Q.; Li, T.S.; Liu, Z.Y.; Wang, J.L.; Qin, Y.; Zhang, X.; Yan, X.W.; et al. The use of anti-inflammatory drugs in the treatment of people with severe coronavirus disease 2019 (COVID-19): The Perspectives of clinical immunologists from China. Clin. Immunol. 2020, 214, 108393. [Google Scholar] [CrossRef]
- Robb, C.T.; Goepp, M.; Rossi, A.G.; Yao, C. Non-steroidal anti-inflammatory drugs, prostaglandins, and COVID-19. Br. J. Pharmacol. 2020, 177, 4899–4920. [Google Scholar] [CrossRef]
- Benveniste, E.N.; Qin, H. Type I interferons as anti-inflammatory mediators. Sci. STKE 2007, 2007, pe70. [Google Scholar] [CrossRef]
- Billiau, A. Anti-inflammatory properties of Type I interferons. Antivir. Res. 2006, 71, 108–116. [Google Scholar] [CrossRef]
- Kovarik, P.; Sauer, I.; Schaljo, B. Molecular mechanisms of the anti-inflammatory functions of interferons. Immunobiology 2008, 212, 895–901. [Google Scholar] [CrossRef] [Green Version]
- Veeranki, S.; Duan, X.; Panchanathan, R.; Liu, H.; Choubey, D. IFI16 protein mediates the anti-inflammatory actions of the type-I interferons through suppression of activation of caspase-1 by inflammasomes. PLoS ONE 2011, 6, e27040. [Google Scholar] [CrossRef] [Green Version]
- Yen, J.H.; Kong, W.; Hooper, K.M.; Emig, F.; Rahbari, K.M.; Kuo, P.C.; Scofield, B.A.; Ganea, D. Differential effects of IFN-β on IL-12, IL-23, and IL-10 expression in TLR-stimulated dendritic cells. J. Leukoc. Biol. 2015, 98, 689–702. [Google Scholar] [CrossRef] [Green Version]
- Tu, Y.F.; Chien, C.S.; Yarmishyn, A.A.; Lin, Y.Y.; Luo, Y.H.; Lin, Y.T.; Lai, W.Y.; Yang, D.M.; Chou, S.J.; Yang, Y.P.; et al. A Review of SARS-CoV-2 and the Ongoing Clinical Trials. Int. J. Mol. Sci. 2020, 21, 2657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, M.; Cao, R.; Zhang, L.; Yang, X.; Liu, J.; Xu, M.; Shi, Z.; Hu, Z.; Zhong, W.; Xiao, G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020, 30, 269–271. [Google Scholar] [CrossRef] [PubMed]
- Cao, B.; Wang, Y.; Wen, D.; Liu, W.; Wang, J.; Fan, G.; Ruan, L.; Song, B.; Cai, Y.; Wei, M.; et al. A Trial of Lopinavir-Ritonavir in Adults Hospitalized with Severe Covid-19. N. Engl. J. Med. 2020, 382, 1787–1799. [Google Scholar] [CrossRef]
- Aricò, E.; Bracci, L.; Castiello, L.; Gessani, S.; Belardelli, F. Are we fully exploiting type I Interferons in today’s fight against COVID-19 pandemic? Cytokine Growth Factor Rev. 2020, 54, 43–50. [Google Scholar] [CrossRef]
- Hung, I.F.-N.; Lung, K.-C.; Tso, E.Y.-K.; Liu, R.; Chung, T.W.-H.; Chu, M.-Y.; Ng, Y.-Y.; Lo, J.; Chan, J.; Tam, A.R. Triple combination of interferon beta-1b, lopinavir–ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: An open-label, randomised, phase 2 trial. Lancet 2020, 395, 1695–1704. [Google Scholar] [CrossRef]
- Rahmani, H.; Davoudi-Monfared, E.; Nourian, A.; Khalili, H.; Hajizadeh, N.; Jalalabadi, N.Z.; Fazeli, M.R.; Ghazaeian, M.; Yekaninejad, M.S. Interferon β-1b in treatment of severe COVID-19: A randomized clinical trial. Int. Immunopharmacol. 2020, 88, 106903. [Google Scholar] [CrossRef]
- Teijaro, J.R. Type I interferons in viral control and immune regulation. Curr. Opin. Virol. 2016, 16, 31–40. [Google Scholar] [CrossRef]
- Mesev, E.V.; LeDesma, R.A.; Ploss, A. Decoding type I and III interferon signalling during viral infection. Nat. Microbiol. 2019, 4, 914–924. [Google Scholar] [CrossRef] [PubMed]
- Lokugamage, K.G.; Hage, A.; de Vries, M.; Valero-Jimenez, A.M.; Schindewolf, C.; Dittmann, M.; Rajsbaum, R.; Menachery, V.D. Type I interferon susceptibility distinguishes SARS-CoV-2 from SARS-CoV. J. Virol. 2020, 94, e01410-20. [Google Scholar] [CrossRef] [PubMed]
- Park, A.; Iwasaki, A. Type I and Type III Interferons–Induction, Signaling, Evasion, and Application to Combat COVID-19. Cell Host Microbe 2020, 27, 870–878. [Google Scholar] [CrossRef] [PubMed]
- Munschauer, F.E., III; Kinkel, R.P. Managing side effects of interferon-beta in patients with relapsing-remitting multiple sclerosis. Clin. Ther. 1997, 19, 883–893. [Google Scholar] [CrossRef]
- Asnis, G.M.; De La Garza, R.; Kohn, S.R.; Reinus, J.F.; Henderson, M.; Shah, J. IFN-induced depression: A role for NSAIDs. Psychopharmacol. Bull. 2003, 37, 29–50. [Google Scholar]
- Ozturk, B. Familial Mediterranean Fever and Multiple Sclerosis Successfully Treated With Interferon Beta-1a: A Case Report. Arch Rheumatol. 2019, 34, 443–446. [Google Scholar] [CrossRef]
- Kötter, I.; Günaydin, I.; Zierhut, M.; Stübiger, N. The Use of Interferon α in Behçet Disease: Review of the Literature. In Seminars in Arthritis and Rheumatism; Elsevier: Amsterdam, The Netherlands, 2004; Volume 33, pp. 320–335. [Google Scholar]
- Guarda, G.; Braun, M.; Staehli, F.; Tardivel, A.; Mattmann, C.; Förster, I.; Farlik, M.; Decker, T.; Du Pasquier, R.A.; Romero, P. Type I interferon inhibits interleukin-1 production and inflammasome activation. Immunity 2011, 34, 213–223. [Google Scholar] [CrossRef] [Green Version]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Choi, Y.-M.; Jeong, H.; Park, U.; Cho, N.-H.; Kim, B.-J. A Hepatitis B Virus-Derived Peptide Can Inhibit Infection of Human Lung Cells with SARS-CoV-2 in a Type-1 Interferon-Dependent Manner. Viruses 2021, 13, 1227. https://0-doi-org.brum.beds.ac.uk/10.3390/v13071227
Choi Y-M, Jeong H, Park U, Cho N-H, Kim B-J. A Hepatitis B Virus-Derived Peptide Can Inhibit Infection of Human Lung Cells with SARS-CoV-2 in a Type-1 Interferon-Dependent Manner. Viruses. 2021; 13(7):1227. https://0-doi-org.brum.beds.ac.uk/10.3390/v13071227
Chicago/Turabian StyleChoi, Yu-Min, Hyein Jeong, Uni Park, Nam-Hyuk Cho, and Bum-Joon Kim. 2021. "A Hepatitis B Virus-Derived Peptide Can Inhibit Infection of Human Lung Cells with SARS-CoV-2 in a Type-1 Interferon-Dependent Manner" Viruses 13, no. 7: 1227. https://0-doi-org.brum.beds.ac.uk/10.3390/v13071227