1. Introduction
Forestry policies rely strongly on the available knowledge about forest resources [
1]. Investing in effective forest assessment and monitoring would help to reduce data gaps and, consequently, to support policy-making processes [
2], such as the design of financial incentives and management of sector trade-offs [
3]. At the European scale, one of the objectives of the Ministerial Conference on the Protection of Forests in Europe [
4] is to update the tools for sustainable monitoring and assessing of forestry. The objective is to support sustainable forest management at the regional, national, or European levels by compiling knowledge about the quantity and quality of the goods and services that are produced and used [
5,
6].
One issue that forestry policy must deal with is fragmented rural ownership. This is one of the most significant obstacles for profitable rural management on a global scale [
7,
8,
9]. Fragmented rural ownership is a very common phenomenon in developed countries [
10], and it is gradually increasing due to the dissociation between forestry and small-scale farming, and to the limited involvement of landowners in forest management [
11]. Small plots account for a large share of forestland. In 2010, Hirsch and Schmithüsen [
12] reported that 61% of all European private forest holdings were less than one hectare.
Crucial information about forest plots and dynamics can be efficiently retrieved through remote sensing techniques [
13,
14,
15]. When the focus is the mapping of forest cover, passive satellite sensors are the most-commonly used due to their high radiometric resolution [
16,
17]. However, the spatial resolution of multispectral sensors from freely-available satellite imagery reaches 15 m in Landsat-8—only in the panchromatic spectral band—[
18] and 10 m in four spectral bands from Sentinel-2 [
19]. When land cover is very fragmented these sensors provide images where several land cover classes might be present over the geographic area that correspond to a single pixel, resulting in pixels with mixed radiometry. Although advanced classification methods are available (machine learning, subpixel analysis, library-based, etc.), remote-sensing-based detection of pure forest patches of less than one hectare remains poorly studied. The reviews on remote sensing applied to forest inventories by White et al. [
20] and Gómez et al. [
21], for instance, refer to areas of at least 120 ha and 1000 ha, respectively. The satellite sensors with fine spatial resolution (Pléiades-1, 2 m, World View-4, 1.2 m) could be an alternative to analyze fragmented landscapes. Conrad et al. [
22] performed a crop classification on fields with areas of a minimum size of 0.05 ha using the 6.5 m-resolution imagery of RapidEye. To map smallholders farming (ranging typically from 0.1 to 0.5 ha), Crespin-Boucaud et al. [
23] used one very high spatial resolution image from the Satellite Pour l’Observation de la Terre (SPOT) 6 satellite, along with open-access data provided by Sentinel-2 and Landsat-8. The main limitation in using of high spatial resolution satellite sensors for large areas lies in their high acquisition costs, which can range from
$12.5 Km
2 to
$22.5 Km
2, or even higher depending on the provider, the type of product, and the level of processing, amongst other factors [
24].
In order to be better able to detect small parcels, the information from different sensors and platforms has been evaluated [
25,
26]. Airborne Light Detection and Ranging (LiDAR) is the sensor that is most-commonly used to capture the structural attributes of a forest stand [
27]. LiDAR systems send laser pulses toward the ground and measure the return time for reflections off vegetation surfaces and the ground [
28]. This measured time, together with the coordinates of the lighted point provided by a geopositioning system, allow a LiDAR system to generate a three-dimensional (3D)-point cloud of the scanned surface. Laser pulses can penetrate the canopy permitting the 3D imaging of the vertical strata of the vegetation. The study by Xu et al. [
29], which focuses on data provided by the 5 m-spatial resolution imagery of RapidEye and by aircraft-LiDAR in radiata pine stands, stands out in the field of LiDAR-based detection of very small, forested areas. A different approach was followed by Palenichka et al. [
30], who developed an algorithm for multi-scale segmentation of forested areas, from a stand level to an individual-tree level. Their source of information was airborne LiDAR data.
LiDAR is also used to estimate structural parameters: tree position, height, canopy shape, and species identification [
31,
32]. Aircraft-LiDAR is often used in the statistical area-based approach (ABA) [
33], where forest attributes of an area of interest are inferred through the combination of field measurements and canopy LiDAR point clouds [
34]. When forest inventories require estimates of structural attributes at stand or sub-stand levels (0.5–50 ha) with relative errors below 10%, ABA-LiDAR methods are inadequate due to the need for data at tree level [
21]. LiDAR acquired from Unmanned Aerial Vehicles (UAVs) can be used in these cases, since it yields a denser point cloud, which greatly facilitates the application of the individual tree detection (ITD) approach [
35]. However, LiDAR data is usually acquired ad hoc in the study area [
36,
37,
38].
A data source that can support the managing of small plots at a regional or national scale is the National LiDAR databases, which are available for an entire national territory. In the case of Spain, this database is LiDAR-Plan Nacional de Ortofotografía Aérea (PNOA) [
39]. Although it was released in 2008 and is updated every 4–5 years, is it still under-exploited in forestry. In fact, in Scopus when ‘PNOA’, ‘LiDAR’, and ‘forestry’ are searched in the title, abstract and keywords, only a single document is listed. This kind of data source allows us to completely avoid fieldwork to acquire LiDAR data. Research on the potential of this low-resolution LiDAR data in the characterization of individual trees remains scarce, especially in agroforestry. Nowadays low-resolution LiDAR point clouds are available for the entire national territory of several countries. There is an increasing demand for research and exploitation of these data [
21,
40]. There is evidence of their potential in the field of agroforestry. For instance, Novero et al. [
41] were able to classify land into four crops in the frame of the Phil-LiDAR program of the Philippines, with a point cloud density of 2 points/m
2. With regard to ad hoc LiDAR data acquisition, Parent et al. [
36] developed an automated algorithm for land cover mapping using low density airborne LiDAR (1.56 points/m
2) and high resolution multispectral imagery. Mohan et al. [
37] detected individual trees in coconut plantations using a LiDAR point cloud of 5 points/m
2. Kathuria et al. [
38] executed individual tree detection as well by using LiDAR with a mean point density of 2 points/m
2.
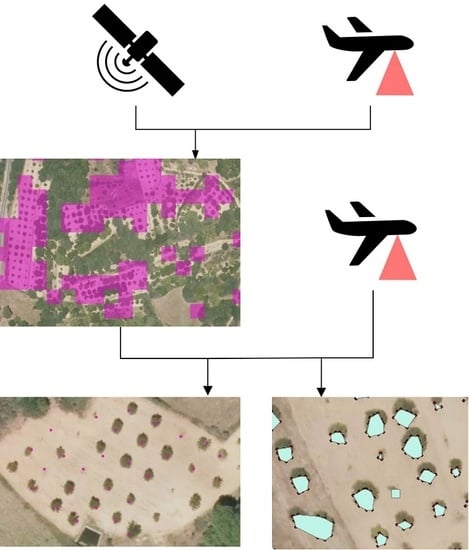
The goal of our research is to explore the potential of the combination of Sentinel-2 satellite images with airborne LiDAR data in the detection, mapping, and characterization of small plantations of trees on a large scale. We used open-access data, specifically data provided by the Copernicus Earth Observation Program and the LiDAR National database [
39]. We adopted the chestnut plantations (
Castanea sativa) of Galicia, a region in northwestern Spain, as a case study. In this region, chestnut plantations have been increasing recently and becoming an important source of income for the area. However, there is no official record of plantations’ locations and characteristics. Sentinel 2 channels and LiDAR derived statistics are processed using supervised classification algorithms in order to locate the small parcels covered by chestnut plantations. Upon location, we performed individual tree detection (ITD) and tree height estimation by applying a local maxima algorithm to a LiDAR derived Canopy Height Model (CHM). We also calculated the crown surface of individual trees by applying a method based on two-dimensional (2D) tree shape reconstruction and canopy segmentation to a projection of the LiDAR point cloud.
4. Discussion
The present study identifies small parcels planted with chestnut trees using LiDAR statistics and Sentinel-2 data with a high overall accuracy. The obtained results reveal that the reduction in the number of variables: a decrease to nine variables (five Sentinel-2 bands and four LiDAR derived statistics) does not significantly compromise the accuracy of the results. Furthermore, the supervised classification algorithm is not a determinant feature in accuracy metrics: it ranges from 92% to 95%. The SVM model is the one that yields the lowest accuracy levels, while the RF provide the highest values. Several other studies report similar results [
64,
75] indicating that the choice of the classifier itself is often of low importance if the data is adequately pre-processed to match the requirements of the classifier [
65].
Despite the high detection accuracy, an overestimation of total chestnut plantation area was observed (
Figure 8). This is due to the peculiarities of the rural environment in the study area. Tree lines and hedges are commonly used to mark parcel boundaries; native species are used, chestnut being one of them. Consequently, the radiometric behavior of these features is often similar to that of plantations. Furthermore, these boundaries also have similar geometric patterns to chestnut plantations: absence of a shrub layer, and analogous tree spacing and canopy density. Isolated trees and forest edges are often erroneously mapped as plantations as well. These are the main causes of the overestimation. However, additional errors, due to the resolution of the source data, often arise when the plantations to be detected are small and irregularly shaped. The result is often a coarse plantation boundary. The area of one square pixel may be mostly an elongated plantation parcel, but it will inevitably include the contiguous surrounding areas, which may exhibit different land usages (
Figure 7).
The particular structure of the plantations analyzed is what makes LiDAR decisive for plantation detection. However, the presented methodology may not be valid if the stand structure changes, especially if there is canopy closure or shrub layer appears. In that case, the presented methodology would need to be modified. Supervised classification using Sentinel-2 data could be enough as there would not be mixed pixels with mixed radiometry (tree-ground). Alonso et al. detected chestnut forests performing a supervised classification on multi-temporal Sentinel-2 data with a previous reduction of the classification area using LiDAR normalized height [
76].
Some other studies have addressed tree plantation detection by combining structural and spectral data obtaining high accuracy levels as well. However, they mainly use high-resolution Satellite data [
77,
78,
79]. The necessity of acquiring high-resolution multispectral data, with its associated costs, could hinder the possibility of performing such studies at a regional or national level.
The ITD process allowed us to estimate the total number of chestnut individuals with a detection accuracy of 90%, which constitutes a suitable value for management purposes. However, some errors were detected. Most of them were related to the overestimation of chestnut plantation area. Since the ITD process is applied to the areas that have been previously mapped as plantations, any errors in the delimitation of their boundaries lead to errors in the counting of chestnut trees. An improvement of the detection method will be needed to avoid that type of errors. Furthermore, branches, cattle, shrubs, and stones are sometimes detected as false positives. However, canopy closure among trees leads to an underestimation of candidates. Despite these deviations, we obtained high accuracy metrics in the ITD and characterization steps. If we compare our results with the ones obtained by Marques et al., who addressed ITD detection in a very similar study case as the one that we present here (chestnut plantation monitoring) but using UAV-captured images [
80], the obtained ITD accuracy is similar (Marques et al. obtained a detection rate of 93.5%). Their approach seams suitable to perform a small-scale study, especially if a National LiDAR database is not available. However, our approach will enable the monitoring of a large area due to the lack of costs and feasibility to have information about the whole territory derived from the use of a National LiDAR dataset.
It should be mentioned that and advantage of the method proposed by Marques et al. [
80] is that the high resolution of UAV-captured images allowed them to obtain better canopy diameter estimations: a RMSE of 0.44 and an R
2 of 0.96. The tree height estimation of Marques et al., however, was not better than the estimation that we obtained using the present methodology: R
2 0.79, RMSE 0.69. This could be due to the inaccuracy of Structure-from-motion (sfM) techniques on ground reconstruction. These errors are propagated into the estimation of individual tree height [
81].
As it was mentioned in the introduction low-density LiDAR is rarely used to characterize individual trees but some studies start to remark the potential of low-density LiDAR for individual tree characterization [
36,
37,
38,
41]. The high accurate metrics obtained in this study are another prove of that potential and encourages to continue studying the potential of low-density ALS for individual tree detection and characterization.
Finally, it should be remarked that the results obtained with the methodology proposed in this study demonstrate that the LiDAR PNOA, together with Sentinel data, enables the creation of cartographic products at a Regional or National level that are useful for forest policy makers and forest managers. This go in line with the conclusions about the opportunity that LiDAR PNOA presents to improve Spanish forest made by Gómez et al. [
21]. This also agrees with White et al. who have also remarked upon the importance of ALS and open-access satellites data to enhance National forest inventories [
20].
5. Conclusions
This study presents a methodology to detect and characterize small plantations of chestnuts. All described processes are based on a combination of low-resolution multispectral data and low-resolution LiDAR point clouds. The multispectral images came from the open-access data from the Sentinel-2 satellite constellation and the LiDAR data came from the open-access database of the Spanish National Mapping Agency.
Using the RF algorithm provided an effective plantation surface estimation, with an overall accuracy of 95.67%. The main limitation observed, which is the coarse delineation of parcel boundaries, is related to the spatial resolution of the satellite images.
ITD through the local maxima algorithm applied to a CHM is an efficient and accurate method for estimating the number of trees in an area and it is powerful enough to locate them. The obtained detection rate and detection accuracy are 96% and 90%, respectively. The limitations in our methods are associated with the previously described error in plantation detection. Our results highlight the need for further research on the potential of low-density LiDAR to assess tree characteristics.
Good forestry policies remain essential for forest managers and are impossible to achieve without thorough knowledge and understanding of resource distribution, extent, and characteristics. This information remains essential in order for policy makers to design policies that are in accordance with the actual situation. The obtained results prove that accurate results useful for forest policy makers and forest managers can be obtained without incurring in high cost products, as it is the case of high-resolution multispectral images or high-resolution LiDAR data. At the same time, the described methodology enables the forest monitoring at a regional or national level, even when the monitoring target consists of very fragmented regions with very small forest parcels scattered around the territory.