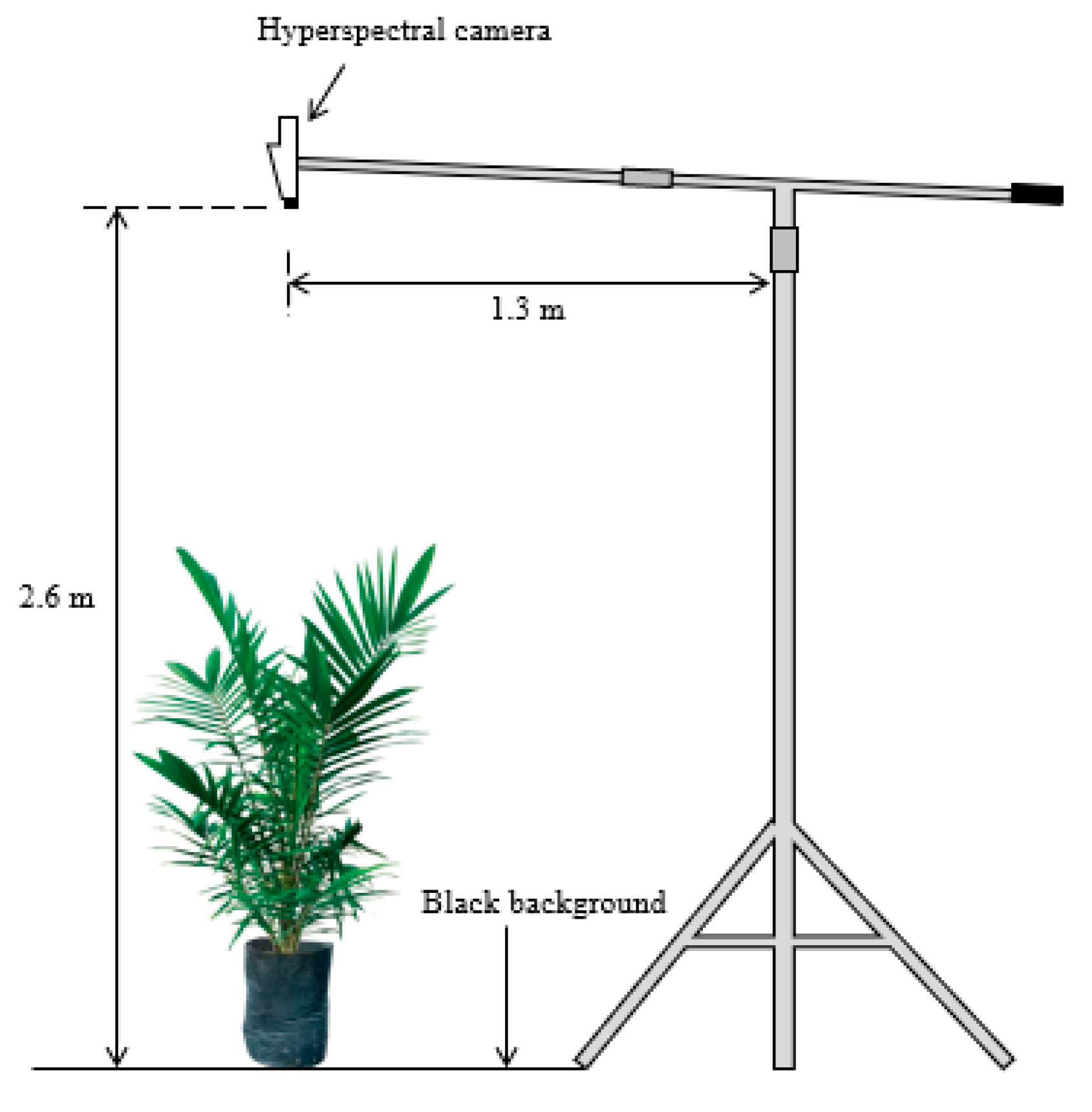
1. Introduction
The production of oil palm in Southeast Asia has been affected by a never-ending case of basal stem rot (BSR) disease. Malaysia has reported annual losses of up to RM 1.5 billion due to this disease, which has made it the most economically devastating disease in the agricultural field. Not only the mature trees, but the oil palm seedlings are also susceptible to the infection whereby the symptoms appear earlier and more severe [
1]. BSR affects a plantation by reducing the number of standing trees as well as the weight of fresh fruit bunches (FFB) [
2]. According to Subagio and Foster [
3], FFB yield decreases by an average of 0.16 tons per hectare for every dead palm or equivalent to 35% when half of the planted trees have died. Based on the BSR incident rate, the total area affected in 2020 was estimated to be 443,440 hectares, equivalent to 65.6 million oil palms, which is worrying if preventative measures are not implemented [
4]. BSR could cause 80% of the affected trees to die [
5]. However, the trees with an infection of less than 20% can still be treated [
6].
G. boninense infection affects the xylem, which causes a major problem in water and nutrient distribution, making the symptoms appear similar to water stress and nutrient deficiency. The earliest visual symptom in seedlings can be seen in the presence of a fruiting body at the bole and followed by partial yellowing of older leaves or mottling of basal fronds to form necrosis, indicating that over 50% of the stem base has been internally damaged [
7]. However, the fruiting body may present or may not before or after the development of foliar symptoms [
5,
8], making visual identification hard, confusing, and often overlooked. In a severe infection, there is no development of new leaves, no increase in height or girth that will consequently lead to growth inhibition. These are all attributed to the inability of the plant to perform normal photosynthesis due to low chlorophyll content and water deficiency [
9]. Despite the visual symptoms,
G. boninense infection can also be detected in the roots and longitudinal sectioning of the infected bole. The longitudinal section usually shows brown discoloration with white mycelium protruding through the root epidermis, indicating upward infection progress within the seedling. Nevertheless, detection through the bole and roots is impractical for large-scale planting areas as the process is labor intensive and causes injury that could lead to tree destruction.
Early detection, timely prevention, and control are indispensable to counter this disease. Nowadays,
G. boninense infection can be detected by various methods. For example, (i) a colorimetric method using ethylenediaminetetraacetic acid (EDTA) [
10], (ii) Ganoderma-selective medium (GSM) [
11], (iii) polyclonal antibodies (PAbs) [
12,
13], (iv) enzyme-linked immunosorbent assay (ELISA) [
14,
15], (v) polymerase chain reaction (PCR) [
16,
17], (vi) an electronic nose (e-nose) device [
18,
19], (vii) microfocus X-ray fluorescence (μXRF) [
6] (viii) tomography images [
20], and (ix) a terrestrial laser scanner (TLS) device [
21,
22,
23]. Nonetheless, these methods are time consuming, impractical for large-scale plantations, and some comprise stem collection and laboratory procedures, which are expensive, tedious, require trained specialists, and appropriate laboratory facilities.
Hyperspectral remote sensing is widely used due to its ability to capture light reflected from plants in narrow and contiguous wavelengths. Every pixel in the image contains a complete spectral reflectance that is well associated with varying degrees and types of stresses [
24,
25,
26,
27,
28,
29,
30]. Healthy plants usually exhibit lower visible reflectance and higher near infrared (NIR) reflectance, whereas unhealthy plants show different spectral patterns depending on the physiology and morphology of the leaves. Advanced data mining analyses can be used to assess vegetation stress; for example, principal component analysis (PCA) [
31,
32], machine learning classification [
29,
30,
33,
34], spectral derivative analysis [
35,
36,
37] and vegetation indices [
38,
39]. Rumpf et al. [
40] combined vegetation indices and SVM to determine healthy and diseased sugar beet leaves caused by
Cercospora beticola. The result showed up to 97% classification accuracy. Nagasubramanian et al. [
41] proved the strength of hyperspectral reflectance over red, green and blue (RGB) data by detecting charcoal rot disease in soybean stem using SVM and obtained 97% and 78% accuracy, respectively.
Nevertheless, the application of hyperspectral imaging to detect BSR in oil palm seedlings has not yet been explored. Instead, it has been applied in several studies of BSR detection in mature oil palm by utilizing an Airborne Imaging Spectrometer for Applications (AISA) imaging system [
42,
43,
44,
45,
46]. These studies were majorly focused on the development of optimal vegetation indices to differentiate between different levels of severity of the BSR disease. However, only a study conducted by Shafri et al. [
44] produced a satisfactorily result with 86% overall accuracy. Alternatively, for oil palm seedlings, other researchers utilized spectroscopy devices to acquire spectral information such as the study performed by Shafri and Anuar [
47], who identified significant bands to distinguish between different severity levels of
G. boninense infection. However, there was poor discrimination between healthy and mildly infected seedlings due to overlapping reflectance spectra between these classes. Similarly, Shafri et al. [
48] also encountered challenges to differentiate between healthy and mildly infected seedlings (inoculated at four months old) after six months of inoculation. Their study used significant bands of first derivative spectra to develop a Maximum Likelihood classification model. The accuracy produced by the model was 82% with a kappa coefficient of 0.73.
Table 1 gives a summary of methods and specific bands used to detect BSR disease in oil palm seedlings and mature trees. As observed, previous studies have been able to present good discrimination accuracy between healthy and infected trees/seedlings with the highest accuracy of 82% and 97% at nursery and plantation level, respectively. Only six out of 20 research studies focused on BSR detection at nursery level using spectroscopy devices with the best method found to be conventional classification, i.e., Maximum Likelihood, and none of the methods used hyperspectral imaging and machine learning techniques. Although the spectroscopy device is capable of detecting early infection of BSR in oil palm seedlings, it has a limitation where the device can only take one reading per time for a small sample point, thus requiring a longer duration of data collection. In contrast, a hyperspectral camera can reduce the time taken for data collection due to its ability to cover large areas in a single imaging session.
The use of machine learning techniques, especially Support Vector Machines (SVMs), has been shown to be beneficial in agriculture applications [
30,
49,
50,
51,
52,
53] and is worth being explored, especially for early detection of BSR such as in studies conducted by Santoso et al. [
54], Santoso et al. [
55] and Khaled et al. [
56]. SVM offers robust analysis and better prediction due to the optimal hyperplane marginal gap between classes. It has been used for classification, regression, and clustering in agricultural studies. The main advantage of SVM is the implementation of the kernel method, which enables higher dimensional separation of non-linear data and improves the computational power of linear learning machines. Kernel functions that are widely used are linear, Gaussian radial basis function (RBF), and polynomial. In addition, this study was carried out to determine the capability of various SVM classifiers to achieve a high degree of accuracy in classifying the different number of bands, features, and frond number for early detection of
G. boninense.
4. Discussion
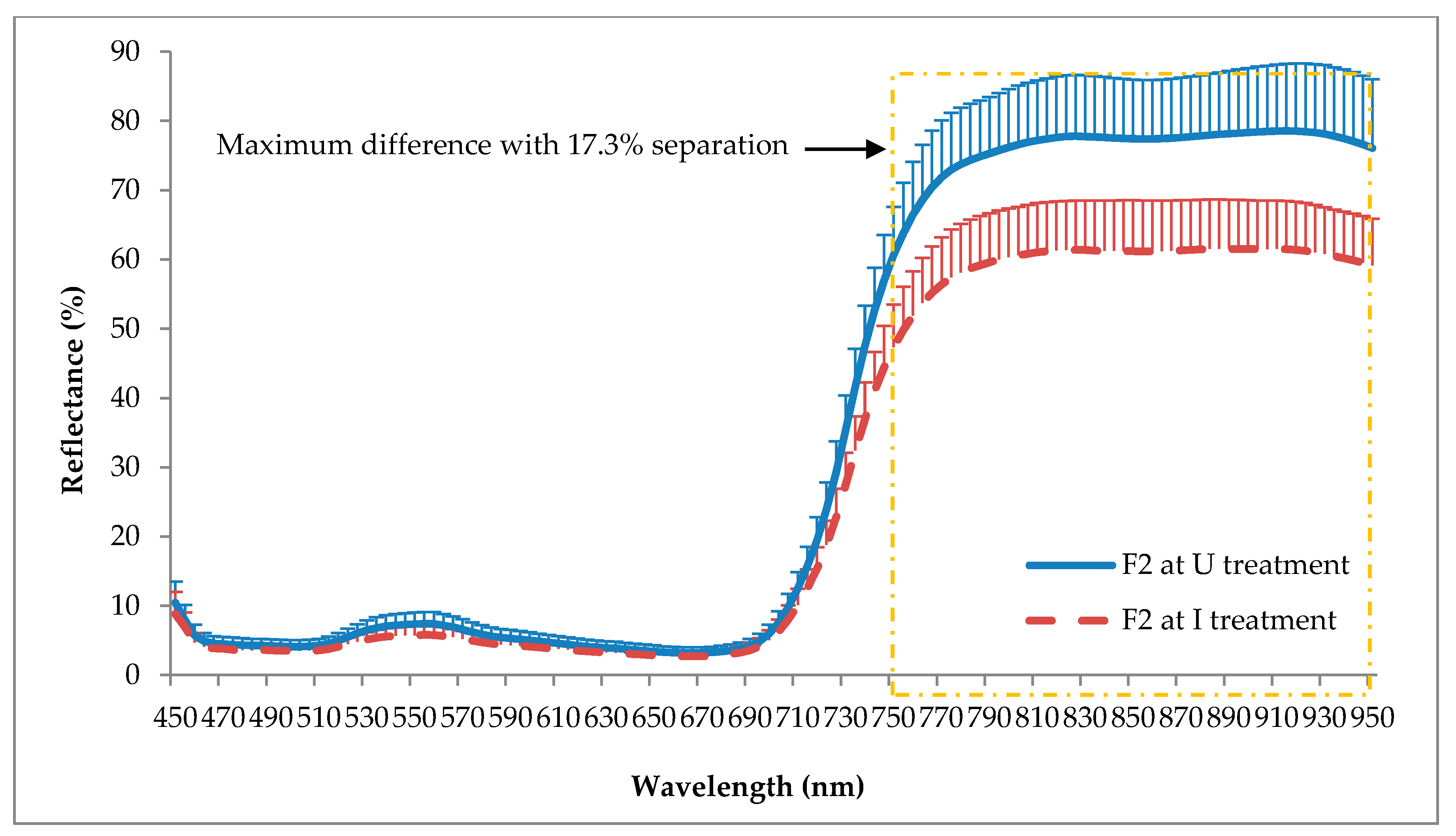
The reflectance pattern generated by I seedlings was typical for diseased plants, with lower reflectance in the NIR spectrum due to the destruction of xylem, which thus reduced the chlorophyll pigments and also caused water deficiency. According to Liaghat et al. [
67], changes of reflectance in the NIR spectrum during a stress period were more evident than changes in the visible spectrum, since NIR could penetrate deeper through the leaf pigments compared to the visible wavelengths. The changes in NIR were due to the rupture of the mesophyll cell wall [
62,
77,
78,
79] which caused higher absorbance and lower reflectance of NIR. This result agreed with the findings by Ausmus and Hilty [
80], where healthy maize dwarf mosaic virus-infected leaves showed significant differences in the NIR range even before the development of the physical symptoms, where the NIR reflectance of healthy leaves was higher than infected leaves.
In this study, the U seedlings reflected a slightly higher light level compared to the I seedlings in the visible range. This pattern was contrary to the spectral signature of healthy plants studied by other researchers that agreed healthy plants normally have lower reflectance than a diseased plant in the visible range, especially for green (520 to 560 nm) due to the higher chlorophyll content of the leaves. Nevertheless, the pattern shown in this study was similar to the study conducted by Shafri et al. [
48] where the healthy seedlings yielded higher reflectance than
G. boninense-infected seedlings in the green wavelengths. Therefore, this reflectance pattern might be a unique spectral signature for oil palm seedlings, since each plant has a specific spectral signature. Furthermore, according to Schmidt and Skidmore [
81], different types of vegetation have spectral reflectance that is statistically significant in various spectral regions.
Although the reflectance patterns of the U and I seedlings of F1 and F2 were almost similar, the U of F2 produced a slightly higher reflectance than the U of F1 throughout the spectrum. This may indicate that the older leaves generated higher reflectance than the younger leaves. This idea was supported by Rapaport et al. [
82] who found increases of NIR reflectance of Cabernet Sauvignon in the second week when the fourth leaf (young) of the control treatment moved to the eighth nodal position (old leaf position) and concluded that age variability mainly influenced the differences in reflectance spectra. Contrarily, Ahmadi et al. [
62] presented average reflectance curves of healthy oil palms where frond 17 (old) produced lower NIR reflectance than frond 9 (young) during the first data collection. However, for the second data collection (8 months later), the NIR reflectance of frond 17 and 9 were almost similar. These reflectance spectra were not consistent with the first data collection.
By using the selected wavelengths of 800 to 950 nm, it was possible to obtain 100% classification accuracy in discriminating healthy and asymptomatic
G. boninense-infected seedlings. The results also confirm that the prediction models developed using F1 generally had the most excellent accuracy of 100% when using 35 and 18 bands as input parameters. In addition, the accuracies attained by the SVM models showed that F12 was able to improve the accuracies of F2, which verified that both fronds could be used to detect the
G. boninense infection in oil palm. This outcome agreed with Shafri et al. [
48] who conducted a Maximum Likelihood classification using a combination of F1 and F2 to determine the health status of oil palm seedlings and achieved a net accuracy of 82% with a kappa coefficient of 0.73. Focusing on the similar type of input data used by Shafri et al. [
48], i.e., F1 and F2, our method gave better accuracy with more than 90% at a different number of bands. Shafri et al. [
48] used 24 significant bands (three green bands, 20 red bands and one NIR band) of first derivative spectra as input parameters. Our method that used the NIR spectrum performed well even at the small number of bands, i.e., nine bands with 94% accuracy. Reducing the number of bands has the advantages of being less complex and more economical. This promising result gave useful information in aerial-view applications such as when applying an unmanned aerial vehicle (UAV) for image acquisition since both fronds can be clearly seen from the top-view image and hence could expedite the detection of the
G. boninense disease.
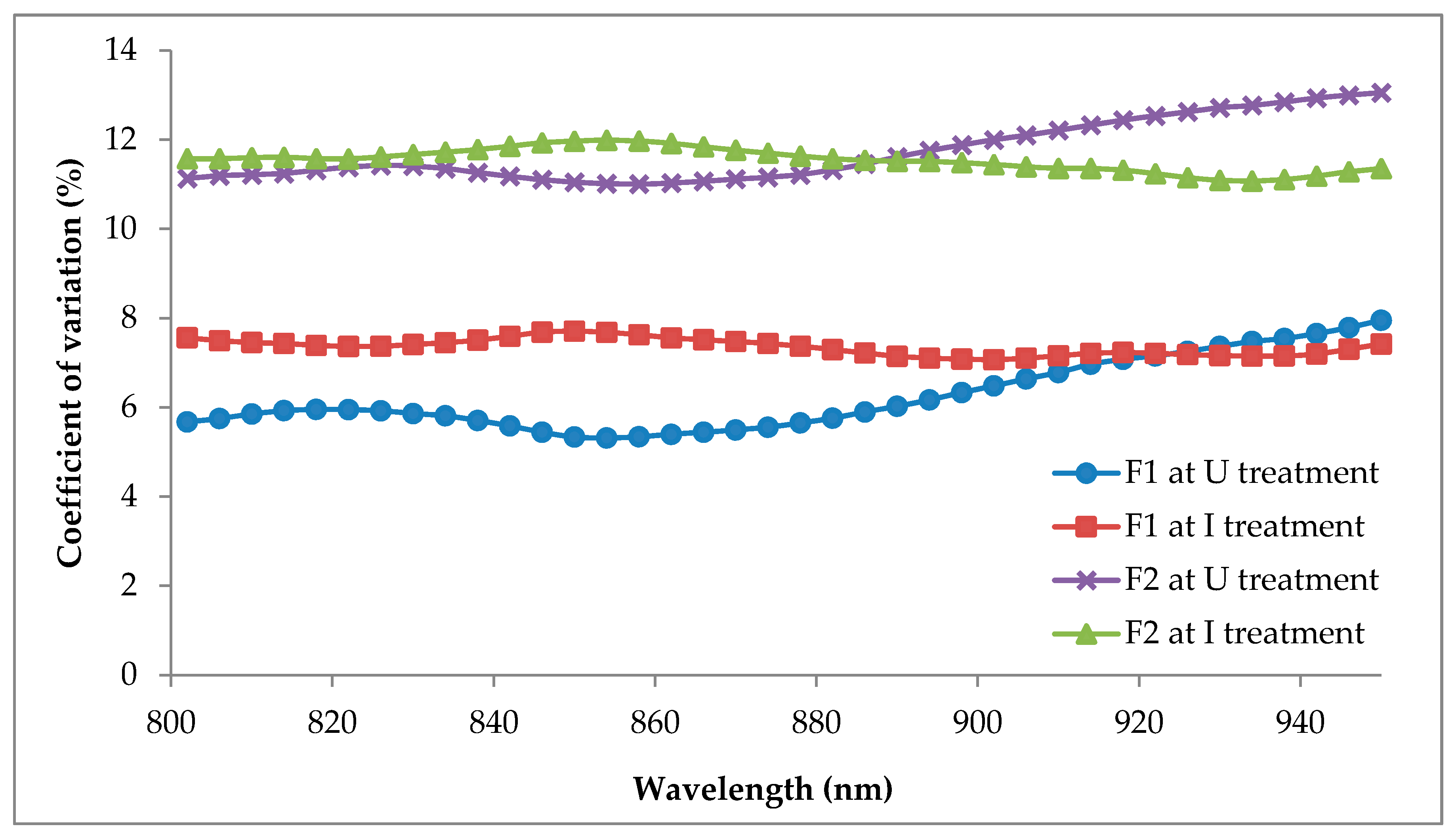
The F1 and F12 classification models produced robust results in contrast to the F2 classification models. For example, several SVM classifiers scored high classification accuracy when using the 35 and 18 (for F1 and F12), 9 and 5 (for F1), and 14 (for F12) bands which suggested that even a small number of input parameters could attain classification accuracies similar to a large number of input parameters. However, it depended on the type of classifiers used. Unlike the F2, the highest classification accuracy was only accomplished by Fine Gaussian SVM using 35 bands. The differences in the number of bands have no significant impact on the accuracy generated by the SVM classifiers. For example, nine bands were able to gain classification accuracies above 90% similar to 35, 18, and 14 bands when processed with F2. The distinct differences between the accuracy of F1 and F2 occurred due to the higher CV of F2, which indicate a higher dispersion of data in F2. In addition, Ahmadi et al. [
62] claimed that younger fronds were more suitable to be used for the early detection of
G. boninense infection since it has a better effect on classification accuracy than the older fronds due to its location on the top part of the crown.
Moreover, the results also present the importance of the kernel method, where Fine Gaussian SVM outperformed Linear SVM with higher classification accuracy in 14 bands of F2, and nine bands of F12, while Medium Gaussian SVM could attain similar accuracies as Linear SVM except when using nine bands of F2. In contrast, Coarse Gaussian SVM obtained less accuracy than the Linear SVM in all bands of F2 but higher in nine bands of F12. Therefore, it showed that the Gaussian RBF classifiers could provide a much better classification than the linear kernel SVM by using the appropriate kernel scale. However, for the polynomial kernel, Quadratic SVM yielded higher classification accuracy than Cubic SVM in F2 and F12, whereas Cubic SVM scored higher classification accuracy than Quadratic SVM in F1. Therefore, this indicated that the performance of SVM-based classification models was highly affected by the different types of data and kernel function, as the kernel effect depended on the data used. For example, F1 data could be optimized to five bands with classification accuracies scoring still above 85% for all SVM classifiers. In comparison, F2 and F12 could only be optimized to 14 bands since the Cubic SVM gave lower accuracy than 85% for both data sets.
5. Conclusions
NIR reflectance showed significant differences between the U and I seedlings. G. boninense infection can be detected at an early stage even though there are no physical symptoms of the disease by using SVM classifiers with a varying number of NIR bands. The reflectance spectra of the F1 frond yielded 100% classification accuracy for all SVM kernel functions using 35 and 18 NIR bands. In contrast, the F2 and F12 fronds achieved the highest classification accuracy of 93% when using Fine Gaussian SVM at 35 NIR bands and 95% when using Gaussian RBF and linear kernel at 35, 18, and 14 NIR bands, respectively. Since F12 produced higher classification accuracy than F2, it could be concluded that F12 would be better used for early detection of G. boninense infection in oil palm when using aerial images because there is no need to separate between F1 and F2 during the pre-processing data stage. Next, it was observed that a high number of bands achieved high classification accuracy while a small number of bands obtained slightly less accuracy. In addition, the optimized Gaussian RBF kernel, i.e., Fine Gaussian SVM could perform excellently compared to the Linear SVM and other classifiers in terms of the classification accuracy produced. However, it depended on the number of bands and fronds used.
For future work, the developed method in this research could be tested in an open environment to confirm its reliability for field application. Even the camera could be calibrated before every image acquisition to avoid error due to illumination. However, careful consideration needs to be taken when dealing with sun angle, shadow and weather conditions. In addition, the inoculation period of oil palm seedlings could be shortened to less 10 months to check the ability to detect the earliest infection of G. boninense. Next, research could also be implemented for different types of oil palm varieties to test their tolerance towards G. boninense infection and its effects on spectral reflectance.