Species-Level Classification and Mapping of a Mangrove Forest Using Random Forest—Utilisation of AVIRIS-NG and Sentinel Data
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Satellite and Airborne Data Processing
2.3. Field Data Collection
2.4. Generation of Vegetation Indices (VIs)
2.5. Mangrove and Non-Forest Mapping: Spectral Angle Mapper (SAM)
2.6. Species-Level Mapping: Random Forest
2.7. Accuracy Assessment
3. Results
3.1. Field Assessment of Species Distribution
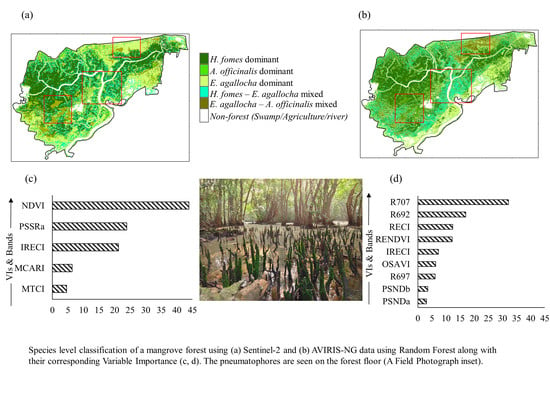
3.2. Species-Level Classification and Mapping Using RF Model
3.3. Importance Variables
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Behera, M.D.; Roy, P.S. Forest Remote Sensing, Biodiversity and Climate Change. Curr. Sci. 2012, 102, 1083–1084. [Google Scholar]
- Kuenzer, C.; Bluemel, A.; Gebhardt, S.; Quoc, T.V.; Dech, S. Remote Sensing of Mangrove Ecosystems: A Review. Remote Sens. 2011, 3, 878–928. [Google Scholar] [CrossRef] [Green Version]
- Behera, M.D. Sundari (H. Fomes)—An Indicator Species of Sundarbans. In Compendium of Biodiversity in Ganga River System; Tare, V., Mathur, R.P., Eds.; LAP Lambert Academic Publishing: Sunnyvale, CA, USA, 2019; pp. 261–278. ISBN 978-613-9-92340-3. [Google Scholar]
- Mickelson, J.G.; Civco, D.L.; Silander, J.A. Delineating Forest Canopy Species in the Northeastern United States Using Multi-Temporal TM Imagery. Photogramm. Eng. Remote Sens. 1998, 64, 891–904. [Google Scholar]
- Hill, R.A.; Wilson, A.K.; George, M.; Hinsley, S.A. Mapping Tree Species in Temperate Deciduous Woodland Using Time-Series Multi-Spectral Data. Appl. Veg. Sci. 2010, 13, 86–99. [Google Scholar] [CrossRef]
- Grabska, E.; Hostert, P.; Pflugmacher, D.; Ostapowicz, K. Forest Stand Species Mapping Using the Sentinel-2 Time Series. Remote Sens. 2019, 11, 1197. [Google Scholar] [CrossRef] [Green Version]
- Martin, M.E.; Newman, S.D.; Aber, J.D.; Congalton, R.G. Determining Forest Species Composition Using High Spectral Resolution Remote Sensing Data. Remote Sens. Environ. 1998, 65, 249–254. [Google Scholar] [CrossRef]
- Wang, T.; Zhang, H.; Lin, H.; Fang, C. Textural-Spectral Feature-Based Species Classification of Mangroves in Mai Po Nature Reserve from Worldview-3 Imagery. Remote Sens. 2016, 8, 24. [Google Scholar] [CrossRef] [Green Version]
- Baloloy, A.B.; Blanco, A.C.; Raymund Rhommel, R.R.C.; Nadaoka, K. Development and Application of a New Mangrove Vegetation Index (MVI) for Rapid and Accurate Mangrove Mapping. ISPRS J. Photogramm. Remote Sens. 2020, 166, 95–117. [Google Scholar] [CrossRef]
- Heenkenda, M.K.; Joyce, K.E.; Maier, S.W.; Bartolo, R. Mangrove Species Identification: Comparing WorldView-2 with Aerial Photographs. Remote Sens. 2014, 6, 6064–6088. [Google Scholar] [CrossRef] [Green Version]
- Heumann, B.W. An Object-Based Classification of Mangroves Using a Hybrid Decision Tree-Support Vector Machine Approach. Remote Sens. 2011, 3, 2440–2460. [Google Scholar] [CrossRef] [Green Version]
- Behera, M.D.; Kushwaha, S.P.S.; Roy, P.S. Forest Vegetation Characterization and Mapping Using IRS-1C Satellite Images in Eastern Himalayan Region. Geocarto Int. 2001, 16, 53–62. [Google Scholar] [CrossRef]
- Sudhakar Reddy, C.; Pattanaik, C.; Murthy, M.S.R. Current Science Association Assessment and Monitoring of Mangroves of Bhitarkanika Wildlife Sanctuary, Orissa, India Using Remote Sensing and GIS Author(s): C. Sudhakar Reddy, Chiranjibi Pattanaik and M.S.R. Murthy Published by: Current Scien. Current Sci. Assoc. 2018, 92, 1409–1415. [Google Scholar]
- Kumar, A.; Stupp, P.; Dahal, S.; Remillard, C.; Bledsoe, R.; Stone, A.; Cameron, C.; Rastogi, G.; Samal, R.; Mishra, D.R. A Multi-Sensor Approach for Assessing Mangrove Biophysical Characteristics in Coastal Odisha, India. Proc. Natl. Acad. Sci. India Sect. A Phys. Sci. 2017, 87, 679–700. [Google Scholar] [CrossRef]
- Gupta, K.; Mukhopadhyay, A.; Giri, S.; Chanda, A.; Datta Majumdar, S.; Samanta, S.; Mitra, D.; Samal, R.N.; Pattnaik, A.K.; Hazra, S. An Index for Discrimination of Mangroves from Non-Mangroves Using LANDSAT 8 OLI Imagery. MethodsX 2018, 5, 1129–1139. [Google Scholar] [CrossRef]
- Ramdani, F.; Rahman, S.; Giri, C. Principal Polar Spectral Indices for Mapping Mangroves Forest in South East Asia: Study Case Indonesia. Int. J. Digit. Earth 2019, 12, 1103–1117. [Google Scholar] [CrossRef]
- Kishore, M.; Kulkarni, S.B. Hyperspectral Imaging Technique for Plant Leaf Identification. In Proceedings of the 2015 International Conference on Emerging Research in Electronics, Computer Science and Technology, ICERECT 2015, Mandya, India, 17–19 December 2015; pp. 209–213. [Google Scholar] [CrossRef]
- Kumar, T.; Panigrahy, S.; Kumar, P.; Parihar, J.S. Classification of Floristic Composition of Mangrove Forests Using Hyperspectral Data: Case Study of Bhitarkanika National Park, India. J. Coast. Conserv. 2013, 17, 121–132. [Google Scholar] [CrossRef]
- Padma, S.; Sanjeevi, S. Jeffries Matusita-Spectral Angle Mapper (JM-SAM) Spectral Matching for Species Level Mapping at Bhitarkanika, Muthupet and Pichavaram Mangroves. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. ISPRS Arch. 2014, 40, 1403–1411. [Google Scholar] [CrossRef] [Green Version]
- Pandey, P.C.; Anand, A.; Srivastava, P.K. Spatial Distribution of Mangrove Forest Species and Biomass Assessment Using Field Inventory and Earth Observation Hyperspectral Data. Biodivers. Conserv. 2019, 28, 2143–2162. [Google Scholar] [CrossRef]
- Chaube, N.R.; Lele, N.; Misra, A.; Murthy, T.V.R.; Manna, S.; Hazra, S.; Panda, M.; Samal, R.N. Mangrove Species Discrimination and Health Assessment Using AVIRIS-NG Hyperspectral Data. Curr. Sci. 2019, 116, 1136–1142. [Google Scholar] [CrossRef]
- Jusoff, K. Individual Mangrove Species Identification and Mapping in Port Klang Using Airborne Hyperspectral Imaging. J. Sustain. Sci. Manag. 2006, 1, 27–36. [Google Scholar]
- Demuro, M.; Chisholm, L. Assessment of Hyperion for Characterizing Mangrove Communities. In Proceedings of the 12th JPL AVIRIS airborne earth science workshop, Pasadena, CA, USA, 12 February 2003; Volume 31, pp. 18–23. [Google Scholar]
- Yang, C.; Everitt, J.H.; Fletcher, R.S.; Jensen, R.R.; Mausel, P.W. Mapping Black Mangrove along the South Texas Gulf Coast Using AISA+ Hyperspectral Imagery. In Proceedings of the Indiana State University—21st Biennial Workshop on Aerial Photography, Videography and High Resolution Digital Imagery for Resource Assessment 2007, Terre Haute, Indiana, 15–17 May 2007; Volume 75, pp. 143–153. [Google Scholar]
- Thomas, N.; Lucas, R.; Bunting, P.; Hardy, A.; Rosenqvist, A.; Simard, M. Distribution and Drivers of Global Mangrove Forest Change, 1996–2010. PLoS ONE 2017, 12, e0179302. [Google Scholar] [CrossRef] [Green Version]
- Cho, M.A.; Debba, P.; Mathieu, R.; Naidoo, L.; Van Aardt, J.; Asner, G.P. Improving Discrimination of Savanna Tree Species through a Multiple-Endmember Spectral Angle Mapper Approach: Canopy-Level Analysis. IEEE Trans. Geosci. Remote Sens. 2010, 48, 4133–4142. [Google Scholar] [CrossRef]
- Pham, T.D.; Bui, D.T.; Yoshino, K.; Le, N.N. Optimized Rule-Based Logistic Model Tree Algorithm for Mapping Mangrove Species Using ALOS PALSAR Imagery and GIS in the Tropical Region. Environ. Earth Sci. 2018, 77, 1–13. [Google Scholar] [CrossRef]
- Wong, F.K.K.; Fung, T. Combining EO-1 Hyperion and Envisat ASAR Data for Mangrove Species Classification in Mai Po Ramsar Site, Hong Kong. Int. J. Remote Sens. 2014, 35, 7828–7856. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, T.; Liu, M.; Jia, M.; Lin, H.; Chu, L.M.; Devlin, A.T. Potential of Combining Optical and Dual Polarimetric SAR Data for Improving Mangrove Species Discrimination Using Rotation Forest. Remote Sens. 2018, 10, 467. [Google Scholar] [CrossRef] [Green Version]
- Cao, J.; Leng, W.; Liu, K.; Liu, L.; He, Z.; Zhu, Y. Object-Based Mangrove Species Classification Using Unmanned Aerial Vehicle Hyperspectral Images and Digital Surface Models. Remote Sens. 2018, 10, 89. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Xu, R. Exploring the Optimal Integration Levels between SAR and Optical Data for Better Urban Land Cover Mapping in the Pearl River Delta. Int. J. Appl. Earth Obs. Geoinf. 2018, 64, 87–95. [Google Scholar] [CrossRef]
- Arasumani, M.; Singh, A.; Bunyan, M.; Robin, V. Testing the Efficacy of Hyperspectral (AVIRIS-NG), Multispectral (Sentinel-2) and Radar (Sentinel-1) Remote Sensing Images to Detect Native and Invasive Non-Native Trees. Biol. Invasions 2021, 1–17. [Google Scholar] [CrossRef]
- Belgiu, M.; Drăgu, L. Random Forest in Remote Sensing: A Review of Applications and Future Directions. ISPRS J. Photogramm. Remote Sens. 2016, 114, 24–31. [Google Scholar] [CrossRef]
- Wang, D.; Wan, B.; Qiu, P.; Su, Y.; Guo, Q.; Wang, R.; Sun, F.; Wu, X. Evaluating the Performance of Sentinel-2, Landsat 8 and Pléiades-1 in Mapping Mangrove Extent and Species. Remote Sens. 2018, 10, 1468. [Google Scholar] [CrossRef] [Green Version]
- Naidoo, L.; Cho, M.A.; Mathieu, R.; Asner, G. Classification of Savanna Tree Species, in the Greater Kruger National Park Region, by Integrating Hyperspectral and LiDAR Data in a Random Forest Data Mining Environment. ISPRS J. Photogramm. Remote Sens. 2012, 69, 167–179. [Google Scholar] [CrossRef]
- Xia, J.; Yokoya, N.; Pham, T.D. Probabilistic Mangrove Species Mapping with Multiple-Source Remote-Sensing Datasets Using Label Distribution Learning in Xuan Thuy National Park, Vietnam. Remote Sens. 2020, 12, 3834. [Google Scholar] [CrossRef]
- Sims, D.A.; Gamon, J.A. Relationships between Leaf Pigment Content and Spectral Reflectance across a Wide Range of Species, Leaf Structures and Developmental Stages. Remote Sens. Environ. 2002, 81, 337–354. [Google Scholar] [CrossRef]
- Horler, D.N.H.; Dockray, M.; Barber, J. The Red Edge of Plant Leaf Reflectance. Int. J. Remote Sens. 1983, 4, 273–288. [Google Scholar] [CrossRef]
- Clevers, J.G.P.W.; Kooistra, L. Using Hyperspectral Remote Sensing Data for Retrieving Canopy Chlorophyll and Nitrogen Content. IEEE J. Sel. Topics Appl. Earth Obs. Remote Sens. 2012, 5, 574–583. [Google Scholar] [CrossRef]
- Zarco-Tejada, P.J.; Hornero, A.; Beck, P.S.A.; Kattenborn, T.; Kempeneers, P.; Hernández-Clemente, R. Chlorophyll Content Estimation in an Open-Canopy Conifer Forest with Sentinel-2A and Hyperspectral Imagery in the Context of Forest Decline. Remote Sens. Environ. 2019, 223, 320–335. [Google Scholar] [CrossRef]
- Varghese, R.; Behera, M. Annual and Seasonal Variations in Gross Primary Productivity across the Agro-Climatic Regions in India. Environ. Monit. Assess. 2019, 191, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Gitelson, A.; Merzlyak, M.N. Spectral Reflectance Changes Associated with Autumn Senescence of Aesculus hippocastanum L. and Acer platanoides L. Leaves. Spectral Features and Relation to Chlorophyll Estimation. J. Plant Physiol. 1994, 143, 286–292. [Google Scholar] [CrossRef]
- Blackburn, G.A. Quantifying Chlorophylls and Carotenoids at Leaf and Canopy Scales: An Evaluation of Some Hyperspectral Approaches. Remote Sens. Environ. 1998, 66, 273–285. [Google Scholar] [CrossRef]
- Dash, J.; Curran, P.J. The MERIS Terrestrial Chlorophyll Index. Int. J. Remote Sens. 2004, 25, 5403–5413. [Google Scholar] [CrossRef]
- Lu, S.; Lu, X.; Zhao, W.; Liu, Y.; Wang, Z.; Omasa, K. Comparing Vegetation Indices for Remote Chlorophyll Measurement of White Poplar and Chinese Elm Leaves with Different Adaxial and Abaxial Surfaces. J. Exp. Bot. 2015, 66, 5625–5637. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Liu, X.; Li, M.; Fang, M.; Chi, W. Neural-Network Model for Estimating Leaf Chlorophyll Concentration in Rice under Stress from Heavy Metals Using Four Spectral Indices. Biosyst. Eng. 2010, 106, 223–233. [Google Scholar] [CrossRef]
- Wu, C.; Niu, Z.; Tang, Q.; Huang, W.; Rivard, B.; Feng, J. Remote Estimation of Gross Primary Production in Wheat Using Chlorophyll-Related Vegetation Indices. Agric. For. Meteorol. 2009, 149, 1015–1021. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Gritz, Y.; Merzlyak, M.N. Relationships between Leaf Chlorophyll Content and Spectral Reflectance and Algorithms for Non-Destructive Chlorophyll Assessment in Higher Plant Leaves. J. Plant Physiol. 2003, 160, 271–282. [Google Scholar] [CrossRef] [PubMed]
- Frampton, W.J.; Dash, J.; Watmough, G.; Milton, E.J. Evaluating the Capabilities of Sentinel-2 for Quantitative Estimation of Biophysical Variables in Vegetation. ISPRS J. Photogramm. Remote Sens. 2013, 82, 83–92. [Google Scholar] [CrossRef] [Green Version]
- Rondeaux, G.; Steven, M.; Baret, F. Optimization of Soil-Adjusted Vegetation Indices. Remote Sens. Environ. 1996, 55, 95–107. [Google Scholar] [CrossRef]
- Hunt, E.R.; Doraiswamy, P.C.; McMurtrey, J.E.; Daughtry, C.S.T.; Perry, E.M.; Akhmedov, B. A Visible Band Index for Remote Sensing Leaf Chlorophyll Content at the Canopy Scale. Int. J. Appl. Earth Obs. Geoinf. 2012, 21, 103–112. [Google Scholar] [CrossRef] [Green Version]
- Rouse, J.; Haas, R.H.; Schell, J.A.; Deering, D.W. Others Monitoring Vegetation Systems in the Great Plains with ERTS. NASA Spec. Publ. 1974, 351, 309. [Google Scholar]
- Daughtry, C.S.; Walthall, C.; Kim, M.; De Colstoun, E.B.; McMurtrey Iii, J. Estimating Corn Leaf Chlorophyll Concentration from Leaf and Canopy Reflectance. Remote Sens. Environ. 2000, 74, 229–239. [Google Scholar] [CrossRef]
- Ghosh, S.; Behera, M. Aboveground Biomass Estimates of Tropical Mangrove Forest Using Sentinel-1 SAR Coherence Data-The Superiority of Deep Learning over a Semi-Empirical Model. Comput. Geosci. 2021, 150, 104737. [Google Scholar] [CrossRef]
- Bhattacharya, B.K.; Green, R.O.; Rao, S.; Saxena, M.; Sharma, S.; Kumar, K.A.; Srinivasulu, P.; Sharma, S.; Dhar, D.; Bandyopadhyay, S.; et al. An Overview of AVIRIS-NG Airborne Hyperspectral Science Campaign over India. Curr. Sci. 2019, 116, 1082–1088. [Google Scholar] [CrossRef]
- Mishra, M.K.; Gupta, A.; John, J.; Shukla, B.P.; Dennison, P.; Srivastava, S.; Kaushik, N.K.; Misra, A.; Dhar, D. Retrieval of Atmospheric Parameters and Data-Processing Algorithms for AVIRIS-NG Indian Campaign Data. Curr. Sci. 2019, 116, 1089–1100. [Google Scholar] [CrossRef]
- Harken, J.; Sugumaran, R. Classification of Iowa Wetlands Using an Airborne Hyperspectral Image: A Comparison of the Spectral Angle Mapper Classifier and an Object-Oriented Approach. Can. J. Remote Sens. 2005, 31, 167–174. [Google Scholar] [CrossRef]
- Na, X.; Zhang, S.; Li, X.; Yu, H.; Liu, C. Improved Land Cover Mapping Using Random Forests Combined with Landsat Thematic Mapper Imagery and Ancillary Geographic Data. Photogramm. Eng. Remote Sens. 2010, 76, 833–840. [Google Scholar] [CrossRef]
- Kulkarni, A.D.; Lowe, B. Random Forest Algorithm for Land Cover Classification. Int. J. Recent Innov. Trends Comput. Commun. 2016, 4, 58–63. [Google Scholar]
- Flores-de-Santiago, F.; Kovacs, J.M.; Wang, J.; Flores-Verdugo, F.; Zhang, C.; González-Farías, F. Examining the Influence of Seasonality, Condition, and Species Composition on Mangrove Leaf Pigment Contents and Laboratory Based Spectroscopy Data. Remote Sens. 2016, 8, 226. [Google Scholar] [CrossRef] [Green Version]
- Ghosh, S.M.; Behera, M.D.; Paramanik, S. Canopy Height Estimation Using Sentinel Series Images through Machine Learning Models in a Mangrove Forest. Remote Sens. 2020, 12, 1519. [Google Scholar] [CrossRef]
- Foody, G.M. Status of Land Cover Classification Accuracy Assessment. Remote Sens. Environ. 2002, 80, 185–201. [Google Scholar] [CrossRef]
- Fang, S.; Gertner, G.; Wang, G.; Anderson, A. The Impact of Misclassification in Land Use Maps in the Prediction of Landscape Dynamics. Landsc. Ecol. 2006, 21, 233–242. [Google Scholar] [CrossRef]
- Foody, G.M. Harshness in Image Classification Accuracy Assessment. Int. J. Remote Sens. 2008, 29, 3137–3158. [Google Scholar] [CrossRef] [Green Version]
- Cohen, J. A Coefficient of Agreement for Nominal Scales. Educ. Psychol. Meas. 1960, 20, 37–46. [Google Scholar] [CrossRef]
- Paramanik, S.; Behera, M.D.; Bhattacharya, B.; Tripathi, S. Evaluation and validation of the modis lai algorithm with digital hemispherical photography at bhitar kanika mangrove forest, india. In Proceedings of the IGARSS 2019 IEEE International Geoscience and Remote Sensing, Yokohama, Japan, 28 July–2 August 2019; pp. 6558–6561. [Google Scholar]
- Green, E.P.; Clark, C.D.; Mumby, P.J.; Edwards, A.J.; Ellis, A.C. Remote Sensing Techniques for Mangrove Mapping. Int. J. Remote Sens. 1998, 19, 935–956. [Google Scholar] [CrossRef]
- Adam, E.; Mutanga, O.; Rugege, D. Multispectral and Hyperspectral Remote Sensing for Identification and Mapping of Wetland Vegetation: A Review. Wetl. Ecol. Manag. 2010, 18, 281–296. [Google Scholar] [CrossRef]
- Millard, K.; Richardson, M. On the Importance of Training Data Sample Selection in Random Forest Image Classification: A Case Study in Peatland Ecosystem Mapping. Remote Sens. 2015, 7, 8489–8515. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Y.; Liu, K.; Liu, L.; Wang, S.; Liu, H. Retrieval of Mangrove Aboveground Biomass at the Individual Species Level with Worldview-2 Images. Remote Sens. 2015, 7, 12192–12214. [Google Scholar] [CrossRef] [Green Version]
- Manna, S.; Raychaudhuri, B. Mapping Distribution of Sundarban Mangroves Using Sentinel-2 Data and New Spectral Metric for Detecting Their Health Condition. Geocarto Int. 2020, 35, 434–452. [Google Scholar] [CrossRef]
- Behera, M.D.; Srivastava, V.K. ERS-1 SAR and Landsat-4 TM Synergism for Forest Cover Studies. Int. J. Geoinformatics 2008, 4, 13–20. [Google Scholar]
- Zhang, C.; Kovacs, J.M.; Liu, Y.; Flores-Verdugo, F.; Flores-de-Santiago, F. Separating Mangrove Species and Conditions Using Laboratory Hyperspectral Data: A Case Study of a Degraded Mangrove Forest of the Mexican Pacific. Remote Sens. 2014, 6, 11673–11688. [Google Scholar] [CrossRef] [Green Version]
- Asner, G.P. Biophysical and Biochemical Sources of Variability in Canopy Reflectance. Remote Sens. Environ. 1998, 64, 234–253. [Google Scholar] [CrossRef]
- Thenkabail, P.S.; Enclona, E.A.; Ashton, M.S.; Van Der Meer, B. Accuracy Assessments of Hyperspectral Waveband Performance for Vegetation Analysis Applications. Remote Sens. Environ. 2004, 91, 354–376. [Google Scholar] [CrossRef]
- Novack, T.; Esch, T.; Kux, H.; Stilla, U. Machine Learning Comparison between WorldView-2 and QuickBird-2-Simulated Imagery Regarding Object-Based Urban Land Cover Classification. Remote Sens. 2011, 3, 2263–2282. [Google Scholar] [CrossRef] [Green Version]





| Sl. No. | Index Name | Formula | Index Range | Interpretation | Reference |
|---|---|---|---|---|---|
| 1 | ReNDVI (Red Edge Normalised Vegetation Index) | (R832 − R717)/(R832 + R717) | −1 to 1 | Inclusion of red edge band provides a good proxy of the chlorophyll content and LAI. | [42,47] |
| 2 | ReCI (Red Edge Chlorophyll Index) | (R832/R717) − 1 | −1 to ∞ | The total chlorophyll content is linearly correlated with the difference between the reciprocal reflectance of green/red edge bands and the NIR band. | [39,48] |
| 3 | IRECI (Inverted Red Edge Chlorophyll Index) | (R783 − R635)/(R705/R740) | 0 to ∞ | Incorporates two red edge bands at wavelength 705 and 740 nm. Least importance given on red band to avoid the saturation. | [49] |
| 4 | (i) PSNDa and (ii) PSNDb (Pigment Specific Normalised Difference for chlorophyll a and b) | (i) (R800 − R680)/(R800 + R680) (ii) (R800 − R635)/(R800 + R635) | −1 to 1 | Chlorophyll a and b were found to be sensitive at wavelengths 680 and 635 nm, respectively. | [43] |
| 5 | OSAVI (Optimized Soil Adjusted Vegetation Index) | (R865 − R660)/(R865 + R660 + 0.16) | −0.86 to +0.86 | Modified SAVI: value of constant (L) was optimized to 0.16 to minimise the background soil reflectance. | [50,51] |
| 6 | NDVI (Normalized Difference Vegetation Index) | (R842 − R665)/(R842 + R665) | −1 to 1 | A measure of the photosynthetic activity and is strongly in correlation with density and vitality of the vegetation. | [52] |
| 7 | MTCI (Meris Terrestrial Chlorophyll Index) | (R740 − R705)/(R705 − R665) | −1 to ∞ | Used for chlorophyll estimation. | [44] |
| 8 | PSSRa (Pigment-Specific Simple Ratio Index) | R783/R665 | 0 to ∞ | To investigate the potential of a range of spectral approaches for quantifying pigments at the scale of the whole plant canopy. | [43] |
| 9 | MCARI (Modified Chlorophyll Absorption Ratio Index) | [(R705 − R665)-0.2 × (R705 − R560) × (R705/R665)] | −∞ to ∞ | To observe the responsiveness to both leaf chlorophyll concentrations and ground reflectance. | [53] |
| Sl. No. | Data Used | Bands/Indices Used | Combination Group | Overall Accuracy (In %) | Kappa Value |
|---|---|---|---|---|---|
| 1 | AVIRIS-NG (Red edge bands) | R692, R697, R702, R707, R707, R717 | Set 1A | 55 | 0.33 |
| 2 | AVIRIS-NG (VIs) | RENDVI (B92, B69), RECI (B92, B69), IRECI (82, 59, 67, 74), PSNDa (86, 53), PSNDb (86, 62), OSAVI (86, 62) | Set 1B | 64 | 0.49 |
| 3 | AVIRIS-NG (3 Red edge bands and 6 VIs) | RENDVI, RECI, IRECI, PSNDa, PSNDb and OSAVI R692, R697, R702 | Set 1D | 70.2 | 0.49 |
| 4 | AVIRIS-NG (6 Red edge bands and 6 VIs) | RENDVI, RECI, IRECI, PSNDa, PSNDb and OSAVI R692, R697, R702, R707, R707, R717 | Set 1C | 67.6 | 0.47 |
| 6 | Sentinel-2 (8 Bands) | R490, R560, R665, R705, R740, R783, R842, R865 | Set 2A | 50 | 0.45 |
| 7 | Sentinel-2 (5 VIs) | MCARI, IRECI, NDVI, PSSRa, MTCI | Set 2B | 74 | 0.61 |
| 8 | Sentinel-2 (5 VIs and 3 bands) | MCARI, IRECI, NDVI, PSSRa, MTCI, B4, B5, B6 | Set 2C | 67 | 0.53 |
| 9 | Sentinel-1 (Dual Pol SAR data) | VH, VV, VH ×VV, ((VV + VV)/2) | Set 3 | 48 | 0.33 |
| 10 | Sentinel-1 and 2 (Bands, VIs, and SAR data) | All above bands, VIs, and SAR data | Set 2 + 3 | 67.5 | 0.54 |
| i. Percentage of Dominance | Heritiera fomes | Excoecaria agallocha | Avicennia officinalis | Heritiera fomes–Excoecaria agallocha | Excoecaria agallocha–Avicennia officinalis | Row Total |
|---|---|---|---|---|---|---|
| >90% | 30 | 22 | 5 | 0 | 0 | 57 |
| >80 to 89% | 19 | 14 | 3 | 0 | 0 | 36 |
| >70 to 79% | 10 | 6 | 6 | 3 | 2 | 27 |
| >60 to 69% | 0 | 0 | 2 | 11 | 9 | 22 |
| >50 to 59% i. Total | 0 59 | 0 42 | 2 18 | 13 27 | 9 20 | 24 166 |
| ii | 30 | 8 | 18 | 37 | 7 | 100 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Behera, M.D.; Barnwal, S.; Paramanik, S.; Das, P.; Bhattyacharya, B.K.; Jagadish, B.; Roy, P.S.; Ghosh, S.M.; Behera, S.K. Species-Level Classification and Mapping of a Mangrove Forest Using Random Forest—Utilisation of AVIRIS-NG and Sentinel Data. Remote Sens. 2021, 13, 2027. https://0-doi-org.brum.beds.ac.uk/10.3390/rs13112027
Behera MD, Barnwal S, Paramanik S, Das P, Bhattyacharya BK, Jagadish B, Roy PS, Ghosh SM, Behera SK. Species-Level Classification and Mapping of a Mangrove Forest Using Random Forest—Utilisation of AVIRIS-NG and Sentinel Data. Remote Sensing. 2021; 13(11):2027. https://0-doi-org.brum.beds.ac.uk/10.3390/rs13112027
Chicago/Turabian StyleBehera, Mukunda Dev, Surbhi Barnwal, Somnath Paramanik, Pulakesh Das, Bimal Kumar Bhattyacharya, Buddolla Jagadish, Parth S. Roy, Sujit Madhab Ghosh, and Soumit Kumar Behera. 2021. "Species-Level Classification and Mapping of a Mangrove Forest Using Random Forest—Utilisation of AVIRIS-NG and Sentinel Data" Remote Sensing 13, no. 11: 2027. https://0-doi-org.brum.beds.ac.uk/10.3390/rs13112027