Predicting Table Beet Root Yield with Multispectral UAS Imagery
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Data Collection
2.3. Data Preprocessing
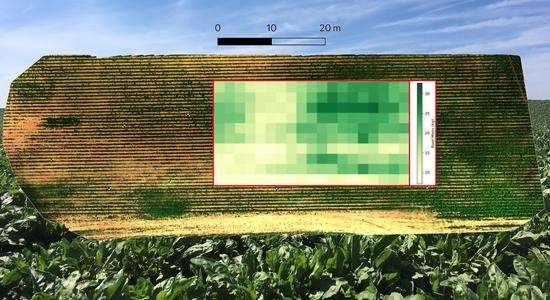
2.4. Canopy Pixel Segmentation
2.5. Feature Choice
2.6. Data Analysis
3. Results
3.1. Table Beet Root Count
3.2. Beet Root Mass 2018
3.3. Beet Root Diameter 2018
3.4. Foliage Mass 2018
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- USDA. National Agricultural Statistics Service 2012 Census of Agriculture; USDA: Washington, DC, USA, 2014.
- Clifford, T.; Howatson, G.; West, D.J.; Stevenson, E.J. The Potential Benefits of Red Beetroot Supplementation in Health and Disease. Nutrients 2015, 7, 2801–2822. [Google Scholar] [CrossRef]
- Gilchrist, M.; Winyard, P.G.; Fulford, J.; Anning, C.; Shore, A.C.; Benjamin, N. Dietary Nitrate Supplementation Improves Reaction Time in Type 2 Diabetes: Development and Application of a Novel Nitrate-Depleted Beetroot Juice Placebo. Nitric Oxide Biol. Chem. 2014, 40, 67–74. [Google Scholar] [CrossRef]
- Hobbs, D.A.; Kaffa, N.; George, T.W.; Methven, L.; Lovegrove, J.A. Blood Pressure-Lowering Effects of Beetroot Juice and Novel Beetroot-Enriched Bread Products in Normotensive Male Subjects. Br. J. Nutr. 2012, 108, 2066–2074. [Google Scholar] [CrossRef] [Green Version]
- Georgiev, V.G.; Weber, J.; Kneschke, E.M.; Denev, P.N.; Bley, T.; Pavlov, A.I. Antioxidant Activity and Phenolic Content of Betalain Extracts from Intact Plants and Hairy Root Cultures of the Red Beetroot Beta Vulgaris cv. Detroit Dark Red. Plant Foods Hum. Nutr. 2010, 65, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Vanhatalo, A.; Bailey, S.J.; Blackwell, J.R.; DiMenna, F.J.; Pavey, T.G.; Wilkerson, D.P.; Benjamin, N.; Winyard, P.G.; Jones, A.M. Acute and Chronic Effects of Dietary Nitrate Supplementation on Blood Pressure and the Physiological Responses to Moderate-Intensity and Incremental Exercise. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2010, 299, 1121–1131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szalaty, M. Physiological Roles and Availability of Betacyanins. Adv. Phytother. 2008, 1, 20–25. [Google Scholar]
- Weiss, M.; Jacob, F.; Duveiller, G. Remote Sensing for Agricultural Applications: A Meta-Review. Remote Sens. Environ. 2020. [Google Scholar] [CrossRef]
- Mulla, D.J. Twenty Five Years of Remote Sensing in Precision Agriculture: Key Advances and Remaining Knowledge Gaps. Biosyst. Eng. 2013, 114, 358–371. [Google Scholar] [CrossRef]
- Pierpaoli, E.; Carli, G.; Pignatti, E.; Canavari, M. Drivers of Precision Agriculture Technologies Adoption: A Literature Review. Procedia Technol. 2013, 8, 61–69. [Google Scholar] [CrossRef] [Green Version]
- Atzberger, C. Advances in Remote Sensing of Agriculture: Context Description, Existing Operational Monitoring Systems and Major Information Needs. Remote Sens. 2013, 5, 949–981. [Google Scholar] [CrossRef] [Green Version]
- Schimmelpfennig, D. Farm Profits and Adoption of Precision Agriculture; USDA: Washington, DC, USA, 2016.
- Stroppiana, D.; Migliazzi, M.; Chiarabini, V.; Crema, A.; Musanti, M.; Franchino, C.; Villa, P. Rice Yield Estimation Using Multispectral Data from UAV: A Preliminary Experiment in Northern Italy. In Proceedings of the 2015 IEEE International Geoscience and Remote Sensing Symposium (IGARSS), Milan, Italy, 26–31 July 2015; pp. 4664–4667. [Google Scholar]
- Duan, B.; Fang, S.; Zhu, R.; Wu, X.; Wang, S.; Gong, Y.; Peng, Y. Remote Estimation of Rice Yield with Unmanned Aerial Vehicle (UAV) Data and Spectral Mixture Analysis. Front. Plant Sci. 2019, 10, 204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maresma, Á.; Lloveras, J.; Martinez-Casasnovas, J.A. Use of Multispectral Airborne Images to Improve In-Season Nitrogen Management, Predict Grain Yield and Estimate Economic Return of Maize in Irrigated High Yielding Environments. Remote Sens. 2018, 10, 543. [Google Scholar] [CrossRef] [Green Version]
- Barzin, R.; Pathak, R.; Lotfi, H.; Varco, J.; Bora, G.C. Use of UAS Multispectral Imagery at Different Physiological Stages for Yield Prediction and Input Resource Optimization in Corn. Remote Sens. 2020, 12, 2392. [Google Scholar] [CrossRef]
- Hassan, M.A.; Yang, M.; Rasheed, A.; Yang, G.; Reynolds, M.; Xia, X.; Xiao, Y.; He, Z. A Rapid Monitoring of NDVI across the Wheat Growth Cycle for Grain Yield Prediction Using a Multi-Spectral UAV Platform. Plant Sci. 2019, 282, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Zheng, H.B.; Xu, X.Q.; He, J.Y.; Ge, X.K.; Yao, X.; Cheng, T.; Zhu, Y.; Cao, W.X.; Tian, Y.C. Predicting Grain Yield in Rice Using Multi-Temporal Vegetation Indices from UAV-Based Multispectral and Digital Imagery. ISPRS J. Photogramm. Remote Sens. 2017, 130, 246–255. [Google Scholar] [CrossRef]
- Olson, D.; Chatterjee, A.; Franzen, D.W.; Day, S.S. Relationship of Drone-Based Vegetation Indices with Corn and Sugarbeet Yields. Agron. J. 2019, 111, 2545–2557. [Google Scholar] [CrossRef]
- Olson, D.; Chatterjee, A.; Franzen, D.W. Can We Select Sugarbeet Harvesting Dates Using Drone-based Vegetation Indices? Agron. J. 2019, 111, 2619–2624. [Google Scholar] [CrossRef]
- Bu, H.; Sharma, L.K.; Denton, A.; Franzen, D.W. Comparison of Satellite Imagery and Ground-Based Active Optical Sensors as Yield Predictors in Sugar Beet, Spring Wheat, Corn, and Sunflower. Agron. J. 2017, 109, 299–308. [Google Scholar] [CrossRef] [Green Version]
- Wanjura, D.F.; Hudspeth, E.B.; Bilbro, J.D. Emergence Time, Seed Quality, and Planting Depth Effects on Yield and Survival of Cotton (Gossypium hirsutum L.). Agron. J. 1969, 61, 63–65. [Google Scholar] [CrossRef]
- Virk, G.; Pilon, C.; Snider, J.L. Impact of First True Leaf Photosynthetic Efficiency on Peanut Plant Growth under Different Early-Season Temperature Conditions. Peanut Sci. 2019, 46, 162–173. [Google Scholar] [CrossRef]
- Raun, W.R.; Solie, J.B.; Johnson, G.V.; Stone, M.L.; Lukina, E.V.; Thomason, W.E.; Schepers, J.S. In-Season Prediction of Potential Grain Yield in Winter Wheat Using Canopy Reflectance. Agron. J. 2001, 93, 131–138. [Google Scholar] [CrossRef] [Green Version]
- Al-Gaadi, K.A.; Hassaballa, A.A.; Tola, E.; Kayad, A.G.; Madugundu, R.; Alblewi, B.; Assiri, F. Prediction of Potato Crop Yield Using Precision Agriculture Techniques. PLoS ONE 2016, 11, e0162219. [Google Scholar] [CrossRef]
- MicaSense RedEdge-M User Manual. Available online: https://support.micasense.com/hc/en-us/articles/115003537673-RedEdge-M-User-Manual-PDF- (accessed on 11 March 2021).
- Pix4D Pix4D Mapper. Available online: https://www.pix4d.com/product/pix4dmapper-photogrammetry-software (accessed on 11 March 2021).
- Mamaghani, B.; Salvaggio, C. Multispectral Sensor Calibration and Characterization for SUAS Remote Sensing. Sensors 2019, 19, 4453. [Google Scholar] [CrossRef] [Green Version]
- Pix4Dmapper Support: Radiometric Corrections. Available online: https://support.pix4d.com/hc/en-us/articles/202559509-Radiometric-corrections (accessed on 18 May 2021).
- MicaSense DLS 2 Integration Guide. Available online: https://support.micasense.com/hc/en-us/articles/360011569434-DLS-2-Integration-Guide (accessed on 18 May 2021).
- L3Harris ENVI. Available online: https://www.l3harrisgeospatial.com/Software-Technology/ENVI (accessed on 11 March 2021).
- Photonics, H. Hyperspectral Software Headwall Photonics Software. Available online: https://www.headwallphotonics.com/software (accessed on 19 May 2021).
- Using ENVI: Atmospheric Correction—Empirical Line Correction. Available online: https://www.l3harrisgeospatial.com/docs/atmosphericcorrection.html#empirical_line_calibration (accessed on 24 May 2021).
- Boggs, T. Spectral Python (SPy). Available online: http://www.spectralpython.net/ (accessed on 11 March 2021).
- Oshigami, S.; Yamaguchi, Y.; Uezato, T.; Momose, A.; Arvelyna, Y.; Kawakami, Y.; Yajima, T.; Miyatake, S.; Nguno, A. Mineralogical Mapping of Southern Namibia by Application of Continuum-Removal MSAM Method to the HyMap Data. Int. J. Remote Sens. 2013, 34, 5282–5295. [Google Scholar] [CrossRef]
- Tichy, L.; Chutry, M.; Botta-Dukát, Z. Semi-Supervised Classification of Vegetation: Preserving the Good Old Units and Searching for New Ones. J. Veg. Sci. 2014, 25, 1504–1512. [Google Scholar] [CrossRef] [Green Version]
- Sader, S.A.; Winne, J.C. RGB-NDVI Colour Composites for Visualizing Forest Change Dynamics. Int. J. Remote Sens. 1992, 13, 3055–3067. [Google Scholar] [CrossRef]
- Gallo, B.C.; Demattê, J.A.M.; Rizzo, R.; Safanelli, J.L.; de Mendes, W.S.; Lepsch, I.F.; Sato, M.V.; Romero, D.J.; Lacerda, M.P.C. Multi-Temporal Satellite Images on Topsoil Attribute Quantification and the Relationship with Soil Classes and Geology. Remote Sens. 2018, 10, 1571. [Google Scholar] [CrossRef]
- Broge, N.H.; Leblanc, E. Comparing Prediction Power and Stability of Broadband and Hyperspectral Vegetation Indices for Estimation of Green Leaf Area Index and Canopy Chlorophyll Density. Remote Sens. Environ. 2001, 76, 156–172. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Merzlyak, M.N.; Lichtenthaler, H.K. Detection of Red Edge Position and Chlorophyll Content by Reflectance Measurements near 700 Nm. J. Plant Physiol. 1996, 148, 501–508. [Google Scholar] [CrossRef]
- Trunk, G.V. A Problem of Dimensionality: A Simple Example. IEEE Trans. Pattern Anal. Mach. Intell. 1979, 3, 306–307. [Google Scholar] [CrossRef] [PubMed]
- Tucker, C.J. Red and Photographic Infrared Linear Combinations for Monitoring Vegetation. Remote Sens. Environ. 1979, 8, 127–150. [Google Scholar] [CrossRef] [Green Version]
- Sripada, R.P. Determining in-Season Nitrogen Requirements for Corn Using Aerial Color-Infrared Photography. Ph.D. Thesis, North Carolina State University, Raleigh, NC, USA, 2005. [Google Scholar]
- Gamon, J.A.; Field, C.B.; Goulden, M.L.; Griffin, K.L.; Hartley, A.E.; Joel, G.; Peñuelas, J.; Valentini, R. Relationships between NDVI, Canopy Structure, and Photosynthesis in Three Californian Vegetation Types. Ecol. Appl. 1995, 5, 28–41. [Google Scholar] [CrossRef] [Green Version]
- Huete, A.; Didan, K.; Miura, T.; Rodriguez, E.P.; Gao, X.; Ferreira, L.G. Overview of the Radiometric and Biophysical Performance of the MODIS Vegetation Indices. Remote Sens. Environ. 2002, 83, 195–213. [Google Scholar] [CrossRef]
- Qi, J.; Chehbouni, A.; Huete, A.R.; Kerr, Y.H.; Sorooshian, S. A Modified Soil Adjusted Vegetation Index. Remote Sens. Environ. 1994, 48, 119–126. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Gritz, Y.; Merzlyak, M.N. Relationships between Leaf Chlorophyll Content and Spectral Reflectance and Algorithms for Non-Destructive Chlorophyll Assessment in Higher Plant Leaves. J. Plant Physiol. 2003, 160, 271–282. [Google Scholar] [CrossRef]
- Haboudane, D.; Miller, J.R.; Pattey, E.; Zarco-Tejada, P.J.; Strachan, I.B. Hyperspectral Vegetation Indices and Novel Algorithms for Predicting Green LAI of Crop Canopies: Modeling and Validation in the Context of Precision Agriculture. Remote Sens. Environ. 2004, 90, 337–352. [Google Scholar] [CrossRef]
- Xiaoqin, W.; Miaomiao, W.; Shaoqiang, W.; Yundong, W. Extraction of Vegetation Information from Visible Unmanned Aerial Vehicle Images. Trans. Chin. Soc. Agric. Eng. 2015, 31, 152–159. [Google Scholar]
- Sims, D.A.; Gamon, J.A. Relationships between Leaf Pigment Content and Spectral Reflectance across a Wide Range of Species, Leaf Structures and Developmental Stages. Remote Sens. Environ. 2002, 81, 337–354. [Google Scholar] [CrossRef]
- Louhaichi, M.; Borman, M.M.; Johnson, D.E. Spatially Located Platform and Aerial Photography for Documentation of Grazing Impacts on Wheat. Geocarto Int. 2001, 16, 65–70. [Google Scholar] [CrossRef]
- Afifi, A.; May, S.; Donatello, R.; Clark, V. Practical Multivariate Analysis, 6th ed.; CRC Press: New York, NY, USA, 2019. [Google Scholar]
- Gu, Y.; Wylie, B.K.; Howard, D.M.; Phuyal, K.P.; Ji, L. NDVI Saturation Adjustment: A New Approach for Improving Cropland Performance Estimates in the Greater Platte River Basin, USA. Ecol. Indic. 2013, 30, 1–6. [Google Scholar] [CrossRef]
- Prabhakara, K.; Hively, W.D.; McCarty, G.W. Evaluating the Relationship between Biomass, Percent Groundcover and Remote Sensing Indices across Six Winter Cover Crop Fields in Maryland, United States. Int. J. Appl. Earth Obs. Geoinf. 2015, 39, 88–102. [Google Scholar] [CrossRef] [Green Version]
- Hassanzadeh, A.; van Aardt, J.; Murphy, S.P.; Pethybridge, S.J. Yield Modeling of Snap Bean Based on Hyperspectral Sensing: A Greenhouse Study. JARS 2020, 14, 024519. [Google Scholar] [CrossRef]
- Pérez-Ortiz, M.; Peña, J.M.; Gutiérrez, P.A.; Torres-Sánchez, J.; Hervás-Martinez, C.; López-Granados, F. A Semi-Supervised System for Weed Mapping in Sunflower Crops Using Unmanned Aerial Vehicles and a Crop Row Detection Method. Appl. Soft Comput. 2015, 37, 533–544. [Google Scholar] [CrossRef]












| Year/Assessment | Ground Truth | UAS Canopy Reflectance | Altitude 1 (m) | GSD 2 (cm) |
|---|---|---|---|---|
| 2018: | ||||
| 1—Emergence | Stand Count (July 5) | 2 Multispectral (July 9) | 14, 27 | 1, 2 |
| 2—Canopy Closing | Stand Count (July 20–22) | 2 Hyperspectral (July 27) | 57, 49 | 3.5, 2.5 |
| 3—Canopy Closed | Stand Count (August 6) | 2 Multispectral (August 9) | 22, 35 | 1.5, 2.5 |
| 4—Harvest | Stand Count (August 20) and Yield Data (August 24) | 2 Multispectral (August 24) | 30, 45 | 2, 3 |
| 2019: | ||||
| 1—Emergence | Stand Count (July 15) | 3 Multispectral (July 16) | 14, 12, 7 | 1, 0.75, 0.5 |
| 2—Canopy Closing | Stand Count (July 24) | 3 Multispectral (July 24) | 14, 12, 7 | 1, 0.75, 0.5 |
| 3—Harvest | Stand Count (August 16) | None | ||
| Band Name | Center Wavelength (nm) | Bandwidth FWHM 1 (nm) |
|---|---|---|
| Blue | 475 | 20 |
| Green | 560 | 20 |
| Red | 668 | 10 |
| Red Edge | 717 | 10 |
| Near Infrared | 840 | 40 |
| Index | Name | Formula |
|---|---|---|
| DVI | Difference Vegetation Index [42] | |
| GDVI | Green Difference Vegetation Index [43] | |
| NDVI | Normalized Difference Vegetation Index [44] | |
| EVI | Enhanced Vegetation Index [45] | |
| MSAVI2 | Modified Soil Adjusted Vegetation Index 2 [46] | |
| GCI | Green Chlorophyll Index [47] | |
| MTVI | Modified Triangular Vegetation Index [48] | |
| VDVI | Visible-Band Difference Vegetation Index [51] | |
| RENDVI | Red Edge Normalized Difference Vegetation Index [50] |
| Band/VI + Area | Area | Band/VI | |||||
|---|---|---|---|---|---|---|---|
| Growth Stage | Best Band/VI | (Roots) | (Roots) | (Roots) | |||
| Emergence | NIR | 0.38 | 22 | 0.23 | 25 | 0.4 | 22 |
| Closing | NIR | 0.03 | 28 | −0.04 | 29 | 0.04 | 28 |
| Closed | VDVI | 0.11 | 27 | −0.02 | 29 | 0.11 | 27 |
| Harvest | NIR | 0.26 | 24 | 0.23 | 25 | 0.14 | 26 |
| Band/VI + Area | Area | Band/VI | |||||
|---|---|---|---|---|---|---|---|
| Growth Stage | Best Band/VI | (kg) | (Roots) | (Roots) | |||
| Emergence | RE | 0.20 | 1.0 | 0.16 | 1.1 | −0.01 | 1.2 |
| Closing | VDVI | 0.37 | 0.9 | 0.36 | 0.9 | −0.03 | 1.2 |
| Closed | MSAVI2 | 0.22 | 1.0 | 0.0 | 1.1 | 0.24 | 1.0 |
| Harvest | R | 0.18 | 1.0 | −0.01 | 1.2 | 0.11 | 1.1 |
| Band/VI + Area | Area | Band/VI | |||||
|---|---|---|---|---|---|---|---|
| Growth Stage | Best Band/VI | (mm) | (mm) | (mm) | |||
| Emergence | NIR | 0.22 | 3.2 | 0.02 | 3.5 | 0.22 | 3.2 |
| Closing | NIR | 0.08 | 3.4 | 0.0 | 3.6 | −0.01 | 3.6 |
| Closed | VDVI | −0.04 | 3.6 | −0.05 | 3.7 | −0.01 | 3.6 |
| Harvest | R | 0.18 | 3.2 | 0.14 | 3.3 | 0.08 | 3.4 |
| Band/VI + Area | Area | Band/VI | |||||
|---|---|---|---|---|---|---|---|
| Growth Stage | Best Band/VI | (g) | (g) | (g) | |||
| Emergence | NDVI | 0.18 | 69 | 0.19 | 68 | 0.09 | 73 |
| Closing | VDVI | 0.26 | 65 | 0.21 | 68 | 0 | 76 |
| Closed | EVI | 0.2 | 68 | 0.03 | 75 | 0.21 | 67.4 |
| Harvest | RENDVI | 0.32 | 63 | 0.13 | 71 | 0.06 | 74 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chancia, R.; van Aardt, J.; Pethybridge, S.; Cross, D.; Henderson, J. Predicting Table Beet Root Yield with Multispectral UAS Imagery. Remote Sens. 2021, 13, 2180. https://0-doi-org.brum.beds.ac.uk/10.3390/rs13112180
Chancia R, van Aardt J, Pethybridge S, Cross D, Henderson J. Predicting Table Beet Root Yield with Multispectral UAS Imagery. Remote Sensing. 2021; 13(11):2180. https://0-doi-org.brum.beds.ac.uk/10.3390/rs13112180
Chicago/Turabian StyleChancia, Robert, Jan van Aardt, Sarah Pethybridge, Daniel Cross, and John Henderson. 2021. "Predicting Table Beet Root Yield with Multispectral UAS Imagery" Remote Sensing 13, no. 11: 2180. https://0-doi-org.brum.beds.ac.uk/10.3390/rs13112180