Single Tree Classification Using Multi-Temporal ALS Data and CIR Imagery in Mixed Old-Growth Forest in Poland
Abstract
:1. Introduction
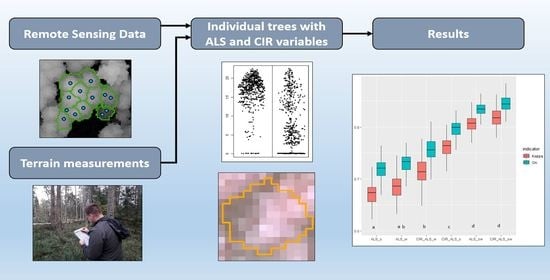
2. Materials and Methods
2.1. Study Area
2.2. ALS Data and CIR Aerial Images
2.3. Field Measurement
2.4. Extracting ALS and CIR Features
2.5. Classification Strategy
2.6. Accuracy Assessment and Statistical Analysis
3. Results
3.1. Classification Results
3.2. Species Classification
3.3. Predictors Importance
4. Discussion
4.1. Classification Results
4.2. Species Classification
4.3. Optimal Data Acquisition
4.4. Predictor’s Importance
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Chaurasia, A.N.; Dave, M.G.; Parmar, R.M.; Bhattacharya, B.; Marpu, P.R.; Singh, A.; Krishnayya, N.S.R. Inferring species diversity and variability over climatic gradient with spectral diversity metrics. Remote Sens. 2020, 12, 2130. [Google Scholar] [CrossRef]
- Ahmed, M.R.; Rahaman, K.R.; Hassan, Q.K. Remote sensing of wildland fire-induced risk assessment at the community level. Sensors 2018, 18, 1570. [Google Scholar] [CrossRef] [Green Version]
- Andrew, M.E.; Wulder, M.A.; Nelson, T.A. Potential contributions of remote sensing to ecosystem service assessments. Prog. Phys. Geogr. 2014, 38, 328–353. [Google Scholar] [CrossRef] [Green Version]
- Yang, D.; Fu, C.S. Mapping regional forest management units: A road-based framework in Southeastern Coastal Plain and Piedmont. For. Ecosyst. 2021, 8, 17. [Google Scholar] [CrossRef]
- Kim, S.; McGaughey, R.J.; Andersen, H.E.; Schreuder, G. Tree species differentiation using intensity data derived from leaf-on and leaf-off airborne laser scanner data. Remote Sens. Environ. 2009, 113, 1575–1586. [Google Scholar] [CrossRef]
- Brandtberg, T. Individual tree-based species classification in high spatial resolution aerial images of forests using fuzzy sets. Fuzzy Sets Syst. 2002, 132, 371–387. [Google Scholar] [CrossRef]
- Clark, M.L.; Roberts, D.A.; Clark, D.B. Hyperspectral discrimination of tropical rain forest tree species at leaf to crown scales. Remote Sens. Environ. 2005, 96, 375–398. [Google Scholar] [CrossRef]
- Immitzer, M.; Atzberger, C.; Koukal, T. Tree species classification with Random forest using very high spatial resolution 8-band worldView-2 satellite data. Remote Sens. 2012, 4, 2661–2693. [Google Scholar] [CrossRef] [Green Version]
- Hycza, T.; Stereńczak, K.; Bałazy, R. Potential use of hyperspectral data to classify forest tree species. N. Z. J. For. Sci. 2018, 48, 18. [Google Scholar] [CrossRef]
- Modzelewska, A.; Kamińska, A.; Fassnacht, F.E.; Stereńczak, K. Multitemporal hyperspectral tree species classification in the Białowieza Forest World Heritage site. For. Int. J. For. Res. 2021, 94, 464–476. [Google Scholar] [CrossRef]
- Heinzel, J.; Koch, B. Investigating multiple data sources for tree species classification in temperate forest and use for single tree delineation. Int. J. Appl. Earth Obs. Geoinf. 2012, 18, 101–110. [Google Scholar] [CrossRef]
- Ghiyamat, A.; Shafri, H.Z.M. A review on hyperspectral remote sensing for homogeneous and heterogeneous forest biodiversity assessment. Int. J. Remote Sens. 2010, 31, 1837–1856. [Google Scholar] [CrossRef]
- Deur, M.; Gašparović, M.; Balenović, I. An Evaluation of Pixel- and Object-Based Tree Species Classification in Mixed Deciduous Forests Using Pansharpened Very High Spatial Resolution Satellite Imagery. Remote Sens. 2021, 13, 1868. [Google Scholar] [CrossRef]
- Clark, M.L.; Clark, D.B.; Roberts, D.A. Small-footprint lidar estimation of sub-canopy elevation and tree height in a tropical rain forest landscape. Remote Sens. Environ. 2004, 91, 68–89. [Google Scholar] [CrossRef]
- Brandtberg, T. Classifying individual tree species under leaf-off and leaf-on conditions using airborne lidar. ISPRS J. Photogramm. Remote Sens. 2007, 61, 325–340. [Google Scholar] [CrossRef]
- Yao, W.; Krzystek, P.; Heurich, M. Tree species classification and estimation of stem volume and DBH based on single tree extraction by exploiting airborne full-waveform LiDAR data. Remote Sens. Environ. 2012, 123, 368–380. [Google Scholar] [CrossRef]
- Shi, Y.; Wang, T.; Skidmore, A.K.; Heurich, M. Important LiDAR metrics for discriminating forest tree species in Central Europe. ISPRS J. Photogramm. Remote Sens. 2018, 137, 163–174. [Google Scholar] [CrossRef]
- Ba, A.; Laslier, M.; Dufour, S.; Hubert-Moy, L. Riparian trees genera identification based on leaf-on/leaf-off airborne laser scanner data and machine learning classifiers in northern France. Int. J. Remote Sens. 2020, 41, 1645–1667. [Google Scholar] [CrossRef]
- Alonzo, M.; Bookhagen, B.; Roberts, D.A. Urban tree species mapping using hyperspectral and lidar data fusion. Remote Sens. Environ. 2014, 148, 70–83. [Google Scholar] [CrossRef]
- Dalponte, M.; Ørka, H.O.; Ene, L.T.; Gobakken, T.; Næsset, E. Tree crown delineation and tree species classification in boreal forests using hyperspectral and ALS data. Remote Sens. Environ. 2014, 140, 306–317. [Google Scholar] [CrossRef]
- Dalponte, M.; Bruzzone, L.; Gianelle, D. Tree species classification in the Southern Alps based on the fusion of very high geometrical resolution multispectral/hyperspectral images and LiDAR data. Remote Sens. Environ. 2012, 123, 258–270. [Google Scholar] [CrossRef]
- Moffiet, T.; Mengersen, K.; Witte, C.; King, R.; Denham, R. Airborne laser scanning: Exploratory data analysis indicates potential variables for classification of individual trees or forest stands according to species. ISPRS J. Photogramm. Remote Sens. 2005, 59, 289–309. [Google Scholar] [CrossRef]
- Korpela, I.; Tokola, T.; Ørka, H.O.; Koskinen, M. Small-footprint discrete-return LIDAR in tree species recognition. In Proceedings of the ISPRS Workshop on High Resolution Earth Imaging for Geospatial Information, Hannover, Germany, 2–5 June 2009. [Google Scholar]
- Hovi, A.; Korhonen, L.; Vauhkonen, J.; Korpela, I. LiDAR waveform features for tree species classification and their sensitivity to tree- and acquisition related parameters. Remote Sens. Environ. 2016, 173, 224–237. [Google Scholar] [CrossRef]
- Kamińska, A.; Lisiewicz, M.; Stereńczak, K.; Kraszewski, B.; Sadkowski, R. Species-related single dead tree detection using multi-temporal ALS data and CIR imagery. Remote Sens. Environ. 2018, 219, 31–43. [Google Scholar] [CrossRef]
- Koenig, K.; Höfle, B. Full-Waveform Airborne Laser Scanning in Vegetation Studies—A Review of Point Cloud and Waveform Features for Tree Species Classification. Forests 2016, 7, 198. [Google Scholar] [CrossRef] [Green Version]
- Harikumar, A.; Paris, C.; Bovolo, F.; Bruzzone, L. A Crown Quantization-Based Approach to Tree-Species Classification Using High-Density Airborne Laser Scanning Data. IEEE Trans. Geosci. Remote Sens. 2020, 59, 4444–4453. [Google Scholar] [CrossRef]
- Holmgren, J.; Persson, Å. Identifying species of individual trees using airborne laser scanner. Remote Sens. Environ. 2004, 90, 415–423. [Google Scholar] [CrossRef]
- Suratno, A.; Seielstad, C.; Queen, L. Tree species identification in mixed coniferous forest using airborne laser scanning. ISPRS J. Photogramm. Remote Sens. 2009, 64, 683–693. [Google Scholar] [CrossRef]
- Korpela, I.; Ørka, H.; Maltamo, M.; Tokola, T.; Hyyppä, J. Tree Species Classification Using Airborne LiDAR—Effects of Stand and Tree Parameters, Downsizing of Training Set, Intensity Normalization, and Sensor Type. Silva Fenn. 2010, 44, 319–339. [Google Scholar] [CrossRef] [Green Version]
- Baltsavias, E.P. Airborne laser scanning: Basic relations and formulas. ISPRS J. Photogramm. Remote Sens. 1999, 54, 199–214. [Google Scholar] [CrossRef]
- Korpela, I.; Ørka, H.O.; Hyyppä, J.; Heikkinen, V.; Tokola, T. Range and AGC normalization in airborne discrete-return LiDAR intensity data for forest canopies. ISPRS J. Photogramm. Remote Sens. 2010, 65, 369–379. [Google Scholar] [CrossRef]
- Ørka, H.O.; Gobakken, T.; Næsset, E.; Ene, L.; Lien, V. Simultaneously acquired airborne laser scanning and multispectral imagery for individual tree species identification. Can. J. Remote Sens. 2012, 38, 125–138. [Google Scholar] [CrossRef]
- Yu, X.; Hyyppä, J.; Litkey, P.; Kaartinen, H.; Vastaranta, M.; Holopainen, M. Single-sensor solution to tree species classification using multispectral airborne laser scanning. Remote Sens. 2017, 9, 108. [Google Scholar] [CrossRef] [Green Version]
- Axelsson, A.; Lindberg, E.; Olsson, H. Exploring Multispectral ALS Data for Tree Species Classification. Remote Sens. 2018, 10, 183. [Google Scholar] [CrossRef] [Green Version]
- Budei, B.C.; St-onge, B.; Hopkinson, C.; Audet, F. Identifying the genus or species of individual trees using a three-wavelength airborne lidar system. Remote Sens. Environ. 2018, 204, 632–647. [Google Scholar] [CrossRef]
- Amiri, N.; Krzystek, P.; Heurich, M.; Skidmore, A. Classification of Tree Species as Well as Standing Dead Trees Using Triple Wavelength ALS in a Temperate Forest. Remote Sens. 2019, 11, 2614. [Google Scholar] [CrossRef] [Green Version]
- Heinzel, J.; Koch, B. Exploring full-waveform LiDAR parameters for tree species classification. Int. J. Appl. Earth Obs. Geoinf. 2011, 13, 152–160. [Google Scholar] [CrossRef]
- Liang, X.; Hyyppä, J.; Matikainen, L. Deciduous-Coniferous Tree Classification Using Difference Between First and Last Pulse Laser Signatures. In Proceedings of the ISPRS Workshop on Laser Scanning 2007 and SilviLaser 2007, Espoo, Finland, 12–14 September 2007; pp. 253–257. [Google Scholar]
- Hollaus, M.; Mücke, W.; Höfle, B.; Dorigo, W.; Pfeifer, N.; Wagner, W.; Bauerhansl, C.; Regner, B. Tree species classification based on full-waveform airborne laser scanning data. In Proceedings of the Silvilaser, College Station, TX, USA, 14–16 October 2009; pp. 54–62. [Google Scholar]
- Ørka, H.O.; Næsset, E.; Bollandsås, O.M. Effects of different sensors and leaf-on and leaf-off canopy conditions on echo distributions and individual tree properties derived from airborne laser scanning. Remote Sens. Environ. 2010, 114, 1445–1461. [Google Scholar] [CrossRef]
- Reitberger, J.; Krzystek, P.; Stilla, U. Analysis of full waveform LIDAR data for the classification of deciduous and coniferous trees. Int. J. Remote Sens. 2008, 29, 1407–1431. [Google Scholar] [CrossRef]
- Laslier, M.; Ba, A.; Hubert-Moy, L.; Dufour, S. Comparison of leaf-on and leaf-off ALS data for mapping riparian tree species. In Proceedings of the SPIE 2017, Warsaw, Poland, 12–15 September 2017. [Google Scholar]
- Yu, X.; Litkey, P.; Hyyppä, J.; Holopainen, M.; Vastaranta, M. Assessment of low density full-waveform airborne laser scanning for individual tree detection and tree species classification. Forests 2014, 5, 1011–1031. [Google Scholar] [CrossRef] [Green Version]
- Sumnall, M.; Hill, R.; Hinsley, S. Comparison of small-footprint discrete return and full waveform airborne lidar data for estimating multiple forest variables. Remote Sens. Environ. 2016, 173, 214–223. [Google Scholar] [CrossRef] [Green Version]
- Stereńczak, K.; Kraszewski, B.; Mielcarek, M.; Kamińska, A.; Lisiewicz, M.; Modzelewska, A.; Sadkowski, R.; Białczak, M.; Piasecka, Ż.; Wilkowska, R. The Białowieża Forest monitoring with the use of remote sensing data. In Zimowa Szkoła Leśna XI Sesja—Zastosowanie Geoinformatyki w Leśnictwie; Instytut Badawczy Leśnictwa: Sękocin Stary, Poland, 2020; pp. 99–107. (In Polish) [Google Scholar]
- Erfanifard, Y.; Stereńczak, K.; Kraszewski, B.; Kamińska, A. Development of a robust canopy height model derived from ALS point clouds for predicting individual crown attributes at the species level. Int. J. Remote Sens. 2018, 39, 9206–9227. [Google Scholar] [CrossRef]
- Stereńczak, K.; Kraszewski, B.; Mielcarek, M.; Piasecka, Ż.; Lisiewicz, M.; Heurich, M. Mapping individual trees with airborne laser scanning data in an European lowland forest using a self-calibration algorithm. Int. J. Appl. Earth Obs. Geoinf. 2020, 93, 102191. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Breiman, L.; Friedman, J.H.; Olshen, R.A.; Stone, C.J. Classification and Regression Trees, 1st ed.; Routledge: Boca Raton, FL, USA, 1984. [Google Scholar]
- Liaw, A.; Wiener, M. Classification and Regression by randomForest. R News 2002, 3, 18–22. [Google Scholar]
- Cohen, J. A Coefficient of Agreement for Nominal Scales. Educ. Psychol. Meas. 1960, 20, 37–46. [Google Scholar] [CrossRef]
- Story, M.; Congalton, R.G. Accuracy Assessment: A User’s Perspective. Photogramm. Eng. Remote Sens. 1986, 52, 397–399. [Google Scholar]
- Bradley, J. Distribution-Free Statistical Tests; Prentice-Hall: Hoboken, NJ, USA, 1968. [Google Scholar]
- Ørka, H.O.; Næsset, E.; Bollandsås, O.M. Utilizing airborne laser intensity for tree species classification. In Proceedings of the ISPRS Workshop on Laser Scanning 2007 and SilviLaser 2007, Espoo, Finland, 12–14 September 2007; pp. 300–304. [Google Scholar]
- Ørka, H.O.; Næsset, E.; Bollandsås, O.M. Classifying species of individual trees by intensity and structure features derived from airborne laser scanner data. Remote Sens. Environ. 2009, 113, 1163–1174. [Google Scholar] [CrossRef]
- Holmgren, J.; Persson, Å.; Söderman, U. Species identification of individual trees by combining high resolution LiDAR data with multi-spectral images. Int. J. Remote Sens. 2008, 29, 1537–1552. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, X. Support vector machines for tree species identification using LiDAR-derived structure and intensity variables. Geocarto Int. 2013, 28, 364–378. [Google Scholar] [CrossRef]
- Lin, Y.; Hyyppä, J. A comprehensive but efficient framework of proposing and validating feature parameters from airborne LiDAR data for tree species classification. Int. J. Appl. Earth Obs. Geoinf. 2016, 46, 45–55. [Google Scholar] [CrossRef]
- You, H.T.; Lei, P.; Li, M.S.; Ruan, F.Q. Forest pecies classification based on three-dimensional coordinate and intensity information of airborne LiDAR data with Random Forest method. In Proceedings of the International Archives of the Photogrammetry, Remote Sensing and Spatial Information Sciences, Guilin, China, 15–17 November 2019; XLII-3/W10, pp. 117–123. [Google Scholar]








| Author | Classes | Species | Study Area |
|---|---|---|---|
| Ba et al. [18] | 8 | oak, alder, poplar, ash, lime, chestnut, willow, beech | Normandy, France |
| Laslier et al. [43] | 8 | ||
| Kim et al. [5] | 2 | Broadleaves (birch, bigleaf maple, elm, magnolia, malus, oak, sorbus, prunus), coniferous (cedar, Douglas fir, larch, pinus, redwood, spruce, western hemlock) | Washington Park Arboretum, Seattle, Washington, USA |
| Shi et al. [17] | 6 | Spruce, beech, fir, birch, maple, rowan | Bavarian Forest National Park, Germany |
| Reitberger et al. [42] | 2 | Coniferous (spruce), deciduous (beech, maple) | |
| Kamińska et al. [25] | 6 | Deciduous (alder, ash, aspen, birch, elm, hornbeam, lime, maple, oak), pine, spruce—divided by dead and alive | Białowieża Forest, Poland |
| Ørka et al. [41] | 2 | Spruce, deciduous (birch, aspen) | Østmarka nature reserve, Norway |
| Species | n | Min (m) | Max (m) | Mean (SD) (m) |
|---|---|---|---|---|
| birch | 117 | 14.4 | 32.8 | 23.5 (4.9) |
| oak | 125 | 13.9 | 36.4 | 28.1 (5.2) |
| hornbeam | 117 | 7.9 | 31.0 | 21.7 (4.5) |
| lime | 98 | 15.4 | 31.8 | 23.6 (4.0) |
| alder | 222 | 10.1 | 33.6 | 25.3 (4.2) |
| pine | 198 | 15.4 | 39.6 | 27.2 (5.2) |
| spruce | 240 | 16.5 | 40.5 | 27.7 (4.9) |
| dead | 113 | 18.4 | 41.4 | 30.5 (5.4) |
| Feature | Description |
|---|---|
| Structural features | |
| Hmean | Arithmetic mean of all normalized heights from the point cloud |
| Hsd | The standard deviation of all normalized heights from the point cloud |
| HCV | The coefficient of variation of all normalized heights from the point cloud |
| Hskew | Skewness of all normalized heights from the point cloud |
| Hkurt | Kurtosis of normalized heights from the point cloud |
| Haad_mean | Average absolute deviation of all normalized heights from the point cloud: mean(abs(X-mean(X)) |
| Haad_median | Median absolute deviation of all normalized heights from the point cloud: median(abs(X-mean(X)) |
| HP10–HP90 | 10th to 90th percentiles of all normalized heights from the point cloud |
| HIQ | Inter-percentile range of all normalized heights from the point cloud: HP75–HP25 |
| Pmean | The ratio of the total number of points above the mean to the total number of all points |
| Pmedian | The ratio of the total number of points above the median to the total number of all points |
| CanRR | Canopy relief ratio of points: (avg(X)-min(X))/(max(X)-min(X)) |
| Additional features | |
| Pfe_all | The proportion of first returns |
| Psingle_all | The proportion of single returns |
| Intensity features | |
| Imean | Mean of intensity values |
| Isd | The standard deviation of intensity values |
| ICV | The coefficient of variation of intensity values |
| Iskew | Skewness of intensity values |
| Ikurt | Kurtosis of intensity values |
| Iaad_mean | Average absolute deviation of intensity values |
| Iaad_median | Median absolute deviation of intensity values |
| IP10–IP90 | 10th to 90th percentiles of intensity values |
| IIQ | Inter-percentile range of intensity values |
| CIR features | |
| NDVI | Normalized differenced vegetation index |
| NIRmean | Mean value of reflectance in the near-infrared band |
| NIRmedian | Median value of reflectance in the near-infrared band |
| NIRsd | The standard deviation of reflectance in the near-infrared band |
| Rmean | Mean value of reflectance in the red band |
| Rmedian | Median value of reflectance in the red band |
| Rsd | The standard deviation of reflectance in the red band |
| Gmean | Mean value of reflectance in the green band |
| Gmedian | Median value of reflectance in the green band |
| Gsd | The standard deviation of reflectance in the green band |
| Symbol | Description |
|---|---|
| ALS | Variant with the usage of ALS point cloud |
| CIR | Variant with the usage of CIR aerial images |
| S | Point cloud from the leaf-on season (summer) |
| W | Point cloud from the leaf-off season (winter) |
| Birch | Alder | Oak | Hornbeam | Lime | Pine | Spruce | Dead | |
|---|---|---|---|---|---|---|---|---|
| ALSS | 0.51(a) | 0.69(a) | 0.58(a) | 0.67(b) | 0.51(b) | 0.84(a) | 0.83(a) | 0.88(a) |
| CIR_ALSS | 0.80(b) | 0.74(ab) | 0.59(ab) | 0.68(b) | 0.53(b) | 0.93(b) | 0.92(b) | 0.98(b) |
| ALSW | 0.53(a) | 0.67(a) | 0.68(bc) | 0.39(a) | 0.34(a) | 0.95(b) | 0.94(b) | 0.94(b) |
| CIR_ALSW | 0.58(a) | 0.69(ab) | 0.76(c) | 0.40(a) | 0.37(ab) | 0.95(b) | 0.95(b) | 0.98(b) |
| ALSSW | 0.86(c) | 0.78(b) | 0.72(c) | 0.66(b) | 0.53(b) | 0.96(b) | 0.95(b) | 0.96(b) |
| CIR_ALSSW | 0.87(c) | 0.78(b) | 0.75(c) | 0.68(b) | 0.55(b) | 0.96(b) | 0.96(b) | 0.98(b) |
| Variants | Predictors |
|---|---|
| ALSS | S_ICV, S_IP90, S_IIQR, S_IP50, S_Iskew |
| ALSW | W_ICV, W_Psingle_all, W_IP40, W_Iskew, W_Pfe_all |
| ALSSW | W_Iskew, W_IP40, W_IP90, W_Psingle_all, S_ICV, W_ICV, S_Iskew, S_IP90 |
| CIR_ALSS | NDVI, S_ICV, Rmean, S_IP90, S_IIQR, S_IP50, S_Iskew |
| CIR_ALSW | W_ICV, W_Psingle_all, W_IP40, NDVI, W_Iskew, W_Pfe_all |
| CIR_ALSSW | W_Iskew, NDVI, S_ICV, W_IP40, W_IP90, W_Psingle_all, W_ICV, S_Iskew, S_IP90 |
| Author | Accuracy | |||
|---|---|---|---|---|
| Classes (n) | Leaf-On | Leaf-Off | Leaf-On and Leaf-Off | |
| Laslier et al. [43] | 8 | OA = 48.1% κ = 0.40 | OA = 45.9% κ = 0.37 | OA = 52.5% κ = 0.45 |
| Kim et al. [5] | 2 | OA = 73.1% | OA = 83.4%, | OA = 90.6% |
| Shi et al. [17] | 6 | OA = 58% κ = 0.47 | OA = 62% κ = 0.51 | OA = 66.5% κ = 0.58 |
| Reitberger et al. [42] | 2 | OA = 85.4% | OA = 95.7% | |
| Kamińska et al. [25] | 6 | OA = 81.4% κ = 0.76 | OA = 87.6% κ = 0.84 | OA = 93.2% κ = 0.91 |
| Ørka et al. [41] | 2 | OA = 0.87 κ = 0.74 | OA = 0.97 κ = 0.94 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kamińska, A.; Lisiewicz, M.; Stereńczak, K. Single Tree Classification Using Multi-Temporal ALS Data and CIR Imagery in Mixed Old-Growth Forest in Poland. Remote Sens. 2021, 13, 5101. https://0-doi-org.brum.beds.ac.uk/10.3390/rs13245101
Kamińska A, Lisiewicz M, Stereńczak K. Single Tree Classification Using Multi-Temporal ALS Data and CIR Imagery in Mixed Old-Growth Forest in Poland. Remote Sensing. 2021; 13(24):5101. https://0-doi-org.brum.beds.ac.uk/10.3390/rs13245101
Chicago/Turabian StyleKamińska, Agnieszka, Maciej Lisiewicz, and Krzysztof Stereńczak. 2021. "Single Tree Classification Using Multi-Temporal ALS Data and CIR Imagery in Mixed Old-Growth Forest in Poland" Remote Sensing 13, no. 24: 5101. https://0-doi-org.brum.beds.ac.uk/10.3390/rs13245101