Critical Assessment of Cocoa Classification with Limited Reference Data: A Study in Côte d’Ivoire and Ghana Using Sentinel-2 and Random Forest Model
Abstract
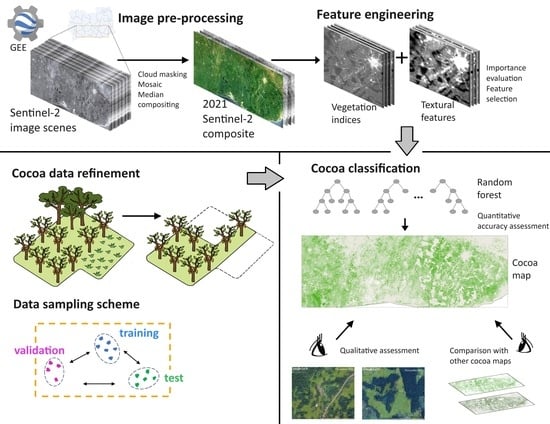
:1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Reference Data
2.2.1. Raw and Refined Cocoa Polygons and Non-Cocoa Polygons Curation
2.2.2. Sampling Design
2.3. Sentinel-2 Imagery Pre-Processing
2.4. Feature Engineering
2.4.1. Vegetation Indices
2.4.2. Textural Features
| Input Data Type | Description | Acronym | Formula | Source |
|---|---|---|---|---|
| Sentinel-2 level-2A reflectance bands | Coastal aerosol | B1 | - | - |
| Blue | B2 | - | - | |
| Green | B3 | - | - | |
| Red | B4 | - | - | |
| Red-edge 1 | B5 | - | - | |
| Red-edge 2 | B6 | - | - | |
| Red-edge 3 | B7 | - | - | |
| Near-infrared | B8 | - | - | |
| Red-edge 4 | B8A | - | - | |
| Water vapor | B9 | - | - | |
| Shortwave infrared 1 | B11 | - | - | |
| Shortwave infrared 2 | B12 | - | - | |
| Vegetation indices | Enhanced vegetation index | EVI | [75] | |
| Green normalised difference vegetation index | GNDVI | [76] | ||
| Normalised difference moisture index | NDMI | [77] | ||
| Normalised difference red edge index | NDRE | [78,79] | ||
| Normalised difference vegetation index | NDVI | [80] | ||
| Plant senescence reflectance index | PSRI | [81] | ||
| Red-edge chlorophyll index | RECI | [72,82] | ||
| Red-edge normalised difference vegetation index | RENDVI | [83] | ||
| Soil-adjusted vegetation index | SAVI | [84] | ||
| Triangular chlorophyll index | TCI | [85] | ||
| Visible atmospherically resistant index | VARI | [86] |
| GLCM Textural Measures | Source | |
|---|---|---|
| Angular second moment (asm) | Inertia (inertia) | [74,87] |
| Cluster prominence (prom) | Information measure of correlation 1 (imcorr1) | |
| Cluster shade (shade) | Information measure of correlation 2 (imcorr2) | |
| Contrast (contrast) | Inverse difference moment (idm) | |
| Correlation (corr) | Sum average (savg) | |
| Difference entropy (dent) | Sum entropy (sen) | |
| Difference variance (dvar) | Sum variance (svar) | |
| Dissimilarity (diss) | Variance (var) | |
| Entropy (ent) | ||
2.4.3. Feature Selection
- TCI, NDVI, GNDVI and B6_savg for the raw dataset;
- NDVI, TCI, B12_savg and B8_savg for the refined dataset.
2.5. Random Forest Classifier
2.6. Quantitative Assessment and Input Configuration
- The four features selected from the importance ranking, with and without the inclusion of the 12 Sentinel-2 bands;
- The 25 features prior to the selection procedure, with and without the inclusion of the 12 Sentinel-2 bands;
- The 12 Sentinel-2 bands solely.
2.7. Qualitative Assessment and Comparison with Previous Cocoa Classification Studies
3. Results
3.1. Quantitative Assessment
3.2. Qualitative Assessment with Visual Inspection
4. Discussion
4.1. Cocoa Data Refinement
4.2. Data Sampling in the Context of Limited Reference Data in Large-Scale Study Area
4.3. Feature Engineering
4.4. Image Compositing
4.5. Qualitative Assessment with Visual Inspection
4.6. Comparison with Other Cocoa Maps and Implications for Reporting on Area Estimation
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A


| s2cloudless Parameters * | Values |
|---|---|
| Cloud filter | 100 |
| Cloud probability threshold | 20 |
| Near-infrared dark threshold | 0.2 |
| Cloud projection distance | 1.5 |
| Buffer | 50 |
| Reference Dataset | Input Data | Non-Cocoa | ||
|---|---|---|---|---|
| UA (%) | PA (%) | F1 (%) | ||
| Refined | 4 features | 79.0 ± 2.2 | 83.3 ± 1.1 | 81.1 ± 1.4 |
| 4 features + 12 Sentinel-2 bands | 83.0 ± 2.9 | 88.3 ± 0.7 | 85.6 ± 1.6 | |
| 25 features | 78.6 ± 2.5 | 88.6 ± 0.6 | 83.3 ± 1.4 | |
| 25 features + 12 Sentinel-2 bands | 81.3 ± 2.6 | 90.1 ± 0.7 | 85.5 ± 1.6 | |
| 12 Sentinel-2 bands | 81.6 ± 3.5 | 88.8 ± 0.8 | 85.0 ± 2.0 | |
| Raw | 4 features | 84.0 ± 2.9 | 79.5 ± 1.1 | 81.6 ± 1.5 |
| 4 features + 12 Sentinel-2 bands | 82.0 ± 3.2 | 85.6 ± 0.8 | 83.8 ± 1.8 | |
| 25 features | 77.0 ± 4.2 | 88.0 ± 0.7 | 82.0 ± 2.5 | |
| 25 features + 12 Sentinel-2 bands | 81.0 ± 3.7 | 89.0 ± 0.9 | 85.0 ± 2.1 | |
| 12 Sentinel-2 bands | 82.0 ± 3.3 | 86.5 ± 0.9 | 84.2 ± 2.0 | |


References
- Joshi, N.; Baumann, M.; Ehammer, A.; Fensholt, R.; Grogan, K.; Hostert, P.; Jepsen, M.R.; Kuemmerle, T.; Meyfroidt, P.; Mitchard, E.T.A.; et al. A Review of the Application of Optical and Radar Remote Sensing Data Fusion to Land Use Mapping and Monitoring. Remote Sens. 2016, 8, 70. [Google Scholar] [CrossRef]
- Mayaux, P.; Pekel, J.-F.; Desclée, B.; Donnay, F.; Lupi, A.; Achard, F.; Clerici, M.; Bodart, C.; Brink, A.; Nasi, R.; et al. State and Evolution of the African Rainforests between 1990 and 2010. Phil. Trans. R. Soc. B 2013, 368, 20120300. [Google Scholar] [CrossRef] [PubMed]
- Reiche, J.; Mullissa, A.; Slagter, B.; Gou, Y.; Tsendbazar, N.-E.; Odongo-Braun, C.; Vollrath, A.; Weisse, M.J.; Stolle, F.; Pickens, A.; et al. Forest Disturbance Alerts for the Congo Basin Using Sentinel-1. Environ. Res. Lett. 2021, 16, 024005. [Google Scholar] [CrossRef]
- Tuanmu, M.-N.; Jetz, W. A Global 1-km Consensus Land-Cover Product for Biodiversity and Ecosystem Modelling. Glob. Ecol. Biogeogr. 2014, 23, 1031–1045. [Google Scholar] [CrossRef]
- Szantoi, Z.; Geller, G.N.; Tsendbazar, N.-E.; See, L.; Griffiths, P.; Fritz, S.; Gong, P.; Herold, M.; Mora, B.; Obregón, A. Addressing the Need for Improved Land Cover Map Products for Policy Support. Environ. Sci. Policy 2020, 112, 28–35. [Google Scholar] [CrossRef]
- Norway’s International Climate and Forest Initiative (NICFI). New Satellite Images to Allow Anyone, Anywhere, to Monitor Tropical Deforestation. Available online: https://www.nicfi.no/current/new-satellite-images-to-allow-anyone-anywhere-to-monitor-tropical-deforestation/ (accessed on 27 September 2023).
- Drusch, M.; Del Bello, U.; Carlier, S.; Colin, O.; Fernandez, V.; Gascon, F.; Hoersch, B.; Isola, C.; Laberinti, P.; Martimort, P.; et al. Sentinel-2: ESA’s Optical High-Resolution Mission for GMES Operational Services. Remote Sens. Environ. 2012, 120, 25–36. [Google Scholar] [CrossRef]
- Torres, R.; Snoeij, P.; Geudtner, D.; Bibby, D.; Davidson, M.; Attema, E.; Potin, P.; Rommen, B.; Floury, N.; Brown, M.; et al. GMES Sentinel-1 Mission. Remote Sens. Environ. 2012, 120, 9–24. [Google Scholar] [CrossRef]
- Gorelick, N.; Hancher, M.; Dixon, M.; Ilyushchenko, S.; Thau, D.; Moore, R. Google Earth Engine: Planetary-Scale Geospatial Analysis for Everyone. Remote Sens. Environ. 2017, 202, 18–27. [Google Scholar] [CrossRef]
- Gong, P.; Liu, H.; Zhang, M.; Li, C.; Wang, J.; Huang, H.; Clinton, N.; Ji, L.; Li, W.; Bai, Y.; et al. Stable Classification with Limited Sample: Transferring a 30-m Resolution Sample Set Collected in 2015 to Mapping 10-m Resolution Global Land Cover in 2017. Sci. Bull. 2019, 64, 370–373. [Google Scholar] [CrossRef] [PubMed]
- Hansen, M.C.; Potapov, P.V.; Moore, R.; Hancher, M.; Turubanova, S.A.; Tyukavina, A.; Thau, D.; Stehman, S.V.; Goetz, S.J.; Loveland, T.R.; et al. High-Resolution Global Maps of 21st-Century Forest Cover Change. Science 2013, 342, 850–853. [Google Scholar] [CrossRef] [PubMed]
- Ge, J.; Qi, J.; Lofgren, B.M.; Moore, N.; Torbick, N.; Olson, J.M. Impacts of Land Use/Cover Classification Accuracy on Regional Climate Simulations. J. Geophys. Res. 2007, 112, D05107. [Google Scholar] [CrossRef]
- McMahon, G. Consequences of Land-Cover Misclassification in Models of Impervious Surface. Photogramm. Eng. Remote Sens. 2007, 73, 1343–1353. [Google Scholar] [CrossRef]
- Li, C.; Wang, J.; Wang, L.; Hu, L.; Gong, P. Comparison of Classification Algorithms and Training Sample Sizes in Urban Land Classification with Landsat Thematic Mapper Imagery. Remote Sens. 2014, 6, 964–983. [Google Scholar] [CrossRef]
- Rogan, J.; Franklin, J.; Stow, D.; Miller, J.; Woodcock, C.; Roberts, D. Mapping Land-Cover Modifications over Large Areas: A Comparison of Machine Learning Algorithms. Remote Sens. Environ. 2008, 112, 2272–2283. [Google Scholar] [CrossRef]
- Jin, H.; Stehman, S.V.; Mountrakis, G. Assessing the Impact of Training Sample Selection on Accuracy of an Urban Classification: A Case Study in Denver, Colorado. Int. J. Remote Sens. 2014, 35, 2067–2081. [Google Scholar] [CrossRef]
- Heydari, S.S.; Mountrakis, G. Effect of Classifier Selection, Reference Sample Size, Reference Class Distribution and Scene Heterogeneity in Per-Pixel Classification Accuracy Using 26 Landsat Sites. Remote Sens. Environ. 2018, 204, 648–658. [Google Scholar] [CrossRef]
- Maxwell, A.E.; Warner, T.A.; Fang, F. Implementation of Machine-Learning Classification in Remote Sensing: An Applied Review. Int. J. Remote Sens. 2018, 39, 2784–2817. [Google Scholar] [CrossRef]
- Ball, J.E.; Anderson, D.T.; Chan, C.S. Comprehensive Survey of Deep Learning in Remote Sensing: Theories, Tools, and Challenges for the Community. J. Appl. Remote Sens. 2017, 11, 042609. [Google Scholar] [CrossRef]
- Ramezan, C.A.; Warner, T.A.; Maxwell, A.E.; Price, B.S. Effects of Training Set Size on Supervised Machine-Learning Land-Cover Classification of Large-Area High-Resolution Remotely Sensed Data. Remote Sens. 2021, 13, 368. [Google Scholar] [CrossRef]
- Copass, C.; Antonova, N.; Kennedy, R. Comparison of Office and Field Techniques for Validating Landscape Change Classification in Pacific Northwest National Parks. Remote Sens. 2019, 11, 3. [Google Scholar] [CrossRef]
- Zhao, Y.; Gong, P.; Yu, L.; Hu, L.; Li, X.; Li, C.; Zhang, H.; Zheng, Y.; Wang, J.; Zhao, Y.; et al. Towards a Common Validation Sample Set for Global Land-Cover Mapping. Int. J. Remote Sens. 2014, 35, 4795–4814. [Google Scholar] [CrossRef]
- Olofsson, P.; Foody, G.M.; Herold, M.; Stehman, S.V.; Woodcock, C.E.; Wulder, M.A. Good Practices for Estimating Area and Assessing Accuracy of Land Change. Remote Sens. Environ. 2014, 148, 42–57. [Google Scholar] [CrossRef]
- Asare, R.; Markussen, B.; Asare, R.A.; Anim-Kwapong, G.; Ræbild, A. On-Farm Cocoa Yields Increase with Canopy Cover of Shade Trees in Two Agro-Ecological Zones in Ghana. Clim. Dev. 2019, 11, 435–445. [Google Scholar] [CrossRef]
- Hainmueller, J.; Hiscox, M.J.; Tampe, M. Sustainable Development for Cocoa Farmers in Ghana; MIT and Harvard University: Cambridge, MA, USA, 2011; p. 66. [Google Scholar]
- Powell, R.L.; Matzke, N.; De Souza, C.; Clark, M.; Numata, I.; Hess, L.L.; Roberts, D.A. Sources of Error in Accuracy Assessment of Thematic Land-Cover Maps in the Brazilian Amazon. Remote Sens. Environ. 2004, 90, 221–234. [Google Scholar] [CrossRef]
- Congalton, R.G.; Green, K. A Practical Look at the Sources of Confusion in Error Matrix Generation. Photogramm. Eng. Remote Sens. 1993, 59, 641–644. [Google Scholar]
- Foody, G.M. Status of Land Cover Classification Accuracy Assessment. Remote Sens. Environ. 2002, 80, 185–201. [Google Scholar] [CrossRef]
- Foody, G.M.; Pal, M.; Rocchini, D.; Garzon-Lopez, C.X.; Bastin, L. The Sensitivity of Mapping Methods to Reference Data Quality: Training Supervised Image Classifications with Imperfect Reference Data. ISPRS Int. J. Geo-Inf. 2016, 5, 199. [Google Scholar] [CrossRef]
- Mellor, A.; Boukir, S.; Haywood, A.; Jones, S. Exploring Issues of Training Data Imbalance and Mislabelling on Random Forest Performance for Large Area Land Cover Classification Using the Ensemble Margin. ISPRS J. Photogramm. Remote Sens. 2015, 105, 155–168. [Google Scholar] [CrossRef]
- Foody, G.M. Ground Reference Data Error and the Mis-Estimation of the Area of Land Cover Change as a Function of its Abundance. Remote Sens. Lett. 2013, 4, 783–792. [Google Scholar] [CrossRef]
- Halladin-Dąbrowska, A.; Kania, A.; Kopeć, D. The t-SNE Algorithm as a Tool to Improve the Quality of Reference Data Used in Accurate Mapping of Heterogeneous Non-Forest Vegetation. Remote Sens. 2020, 12, 39. [Google Scholar] [CrossRef]
- Descals, A.; Wich, S.; Meijaard, E.; Gaveau, D.L.A.; Peedell, S.; Szantoi, Z. High-Resolution Global Map of Smallholder and Industrial Closed-Canopy Oil Palm Plantations. Earth Syst. Sci. Data 2021, 13, 1211–1231. [Google Scholar] [CrossRef]
- Kalischek, N.; Lang, N.; Renier, C.; Daudt, R.; Addoah, T.; Thompson, W.; Blaser-Hart, W.; Garrett, R.; Wegner, J. Satellite-Based High-Resolution Maps of Cocoa Planted Area for Côte d’Ivoire and Ghana. arXiv 2022, arXiv:2206.06119. [Google Scholar] [CrossRef]
- Maskell, G.; Chemura, A.; Nguyen, H.; Gornott, C.; Mondal, P. Integration of Sentinel Optical and Radar Data for Mapping Smallholder Coffee Production Systems in Vietnam. Remote Sens. Environ. 2021, 266, 112709. [Google Scholar] [CrossRef]
- Somarriba, E.; López-Sampson, A. Coffee and Cocoa Agroforestry Systems: Pathways to Deforestation, Reforestation, and Tree Cover Change; The World Bank: Washington, DC, USA, 2018; p. 50. [Google Scholar]
- Asare, R.; Afari-Sefa, V.; Osei-Owusu, Y.; Pabi, O. Cocoa Agroforestry for Increasing Forest Connectivity in a Fragmented Landscape in Ghana. Agroforest. Syst. 2014, 88, 1143–1156. [Google Scholar] [CrossRef]
- Barima, Y.; Kouakou, A.; Bamba, I.; Yao Charles, S.; Godron, M.; Andrieu, J.; Bogaert, J. Cocoa Crops are Destroying the Forest Reserves of the Classified Forest of Haut-Sassandra (Ivory Coast). Glob. Ecol. Conserv. 2016, 8, 85–98. [Google Scholar] [CrossRef]
- Cocoa and Forests Initiative. Cocoa and Forests Initiative. Available online: https://www.idhsustainabletrade.com/initiative/cocoa-and-forests/ (accessed on 27 September 2023).
- Mondelēz International Cocoa Life. Progress Blog. Available online: https://www.cocoalife.org/progress (accessed on 27 September 2023).
- Nestlé Cocoa Plan. Towards Forest Positive Cocoa Progress Report 2023. Available online: https://www.nestlecocoaplan.com/article-towards-forest-positive-cocoa-0 (accessed on 27 September 2023).
- The European Parliament; The Council of the European Union. Regulation (EU) 2023/1115 of the European Parliament and of the Council of 31 May 2023 on the Making Available on the Union Market and the Export from the Union of Certain Commodities and Products Associated with Deforestation and Forest Degradation and Repealing Regulation (EU) No 995/2010; European Union: Brussels, Belgium, 2023; pp. 206–247.
- Ashiagbor, G.; Forkuo, E.K.; Asante, W.A.; Acheampong, E.; Quaye-Ballard, J.A.; Boamah, P.; Mohammed, Y.; Foli, E. Pixel-Based and Object-Oriented Approaches in Segregating Cocoa from Forest in the Juabeso-Bia Landscape of Ghana. Remote Sens. Appl. Soc. Environ. 2020, 19, 100349. [Google Scholar] [CrossRef]
- Sassen, M.; Van Soesbergen, A.; Arnell, A.P.; Scott, E. Patterns of (Future) Environmental Risks from Cocoa Expansion and Intensification in West Africa Call for Context Specific Responses. Land Use Policy 2022, 119, 106142. [Google Scholar] [CrossRef]
- Abu, I.-O.; Szantoi, Z.; Brink, A.; Robuchon, M.; Thiel, M. Detecting Cocoa Plantations in Côte d’Ivoire and Ghana and their Implications on Protected Areas. Ecol. Indic. 2021, 129, 107863. [Google Scholar] [CrossRef] [PubMed]
- Asubonteng, K.; Pfeffer, K.; Ros-Tonen, M.; Verbesselt, J.; Baud, I. Effects of Tree-Crop Farming on Land-Cover Transitions in a Mosaic Landscape in the Eastern Region of Ghana. Environ. Manag. 2018, 62, 529–547. [Google Scholar] [CrossRef]
- Benefoh, D.T.; Villamor, G.B.; Van Noordwijk, M.; Borgemeister, C.; Asante, W.A.; Asubonteng, K.O. Assessing Land-Use Typologies and Change Intensities in a Structurally Complex Ghanaian Cocoa Landscape. Appl. Geogr. 2018, 99, 109–119. [Google Scholar] [CrossRef]
- Numbisi, F.N.; Van Coillie, F.M.B.; De Wulf, R. Delineation of Cocoa Agroforests Using Multiseason Sentinel-1 SAR Images: A Low Grey Level Range Reduces Uncertainties in GLCM Texture-Based Mapping. ISPRS Int. J. Geo-Inf. 2019, 8, 179. [Google Scholar] [CrossRef]
- Läderach, P.; Martinez Valle, A.; Schroth, G.; Castro, N. Predicting the Future Climatic Suitability for Cocoa Farming of the World’s Leading Producer Countries, Ghana and Côte d’Ivoire. Clim. Chang. 2013, 119, 841–854. [Google Scholar] [CrossRef]
- Schroth, G.; Läderach, P.; Martinez Valle, A.; Bunn, C. From Site-Level to Regional Adaptation Planning for Tropical Commodities: Cocoa in West Africa. Mitig. Adapt. Strateg. Glob. Chang. 2017, 22, 903–927. [Google Scholar] [CrossRef]
- FAO; ICRISAT; CIAT. Climate-Smart Agriculture in Côte d’Ivoire. CSA Country Profiles for Africa Series; FAO: Rome, Italy, 2018; p. 23. [Google Scholar]
- Ministry of Food and Agriculture (MOFA); Statistics, Research and Information Directorate (SRID). Agriculture in Ghana: Facts and Figures 2019; MOFA: Accra, Ghana, 2020.
- Forestry Commission. Ghana REDD+ Strategy 2016–2035; Forestry Commission: Accra, Ghana, 2016; p. 101.
- FAO. State of the World’s Forests 2016. Forests and Agriculture: Land-Use Challenges and Opportunities; FAO: Rome, Italy, 2016. [Google Scholar]
- Ruf, F.O. The Myth of Complex Cocoa Agroforests: The Case of Ghana. Hum. Ecol. 2011, 39, 373–388. [Google Scholar] [CrossRef]
- Duguma, B.; Gockowski, J.; Bakala, J. Smallholder Cacao (Theobroma Cacao Linn.) Cultivation in Agroforestry Systems of West and Central Africa: Challenges and Opportunities. Agrofor. Syst. 2001, 51, 177–188. [Google Scholar] [CrossRef]
- Laven, A.; Bymolt, R.; Tyszler, M. Demystifying the Cocoa Sector in Ghana and Côte d’Ivoire; The Royal Tropical Institute (KIT): Amsterdam, The Netherlands, 2018. [Google Scholar]
- Roth, M.; Antwi, Y.A.; O’Sullivan, R. Land and Natural Resource Governance and Tenure for Enabling Sustainable Cocoa Cultivation in Ghana; USAID Tenure and Global Climate Change Program: Washington, DC, USA, 2017.
- Ploton, P.; Mortier, F.; Réjou-Méchain, M.; Barbier, N.; Picard, N.; Rossi, V.; Dormann, C.; Cornu, G.; Viennois, G.; Bayol, N.; et al. Spatial Validation Reveals Poor Predictive Performance of Large-Scale Ecological Mapping Models. Nat. Commun. 2020, 11, 4540. [Google Scholar] [CrossRef] [PubMed]
- Roberts, D.R.; Bahn, V.; Ciuti, S.; Boyce, M.S.; Elith, J.; Guillera-Arroita, G.; Hauenstein, S.; Lahoz-Monfort, J.J.; Schröder, B.; Thuiller, W.; et al. Cross-Validation Strategies for Data with Temporal, Spatial, Hierarchical, or Phylogenetic Structure. Ecography 2017, 40, 913–929. [Google Scholar] [CrossRef]
- ESA. Sentinel-2 User Handbook, 2nd ed.; European Space Agency: Paris, France, 2015.
- Simonetti, D.; Pimple, U.; Langner, A.; Marelli, A. Pan-Tropical Sentinel-2 Cloud-Free Annual Composite Datasets. Data Brief 2021, 39, 107488. [Google Scholar] [CrossRef]
- Zupanc, A. Improving Cloud Detection with Machine Learning. Available online: https://medium.com/sentinel-hub/improving-cloud-detection-with-machine-learning-c09dc5d7cf13 (accessed on 15 September 2023).
- Google Earth Engine. Sentinel-2: Cloud Probability. Available online: https://developers.google.com/earth-engine/datasets/catalog/COPERNICUS_S2_CLOUD_PROBABILITY (accessed on 15 September 2023).
- Skakun, S.; Wevers, J.; Brockmann, C.; Doxani, G.; Aleksandrov, M.; Batič, M.; Frantz, D.; Gascon, F.; Gómez-Chova, L.; Hagolle, O.; et al. Cloud Mask Intercomparison eXercise (CMIX): An Evaluation of Cloud Masking Algorithms for Landsat 8 and Sentinel-2. Remote Sens. Environ. 2022, 274, 112990. [Google Scholar] [CrossRef]
- The Earth Engine Community Authors. Sentinel-2 Cloud Masking with s2cloudless. Available online: https://github.com/google/earthengine-community/blob/master/tutorials/sentinel-2-s2cloudless/index.ipynb (accessed on 15 September 2023).
- Batista, J.E.; Rodrigues, N.M.; Cabral, A.I.R.; Vasconcelos, M.J.P.; Venturieri, A.; Silva, L.G.T.; Silva, S. Optical Time Series for the Separation of Land Cover Types with Similar Spectral Signatures: Cocoa Agroforest and Forest. Int. J. Remote Sens. 2022, 43, 3298–3319. [Google Scholar] [CrossRef]
- Aguilar, R.; Zurita-Milla, R.; Izquierdo-Verdiguier, E.; De By, R.A. A Cloud-Based Multi-Temporal Ensemble Classifier to Map Smallholder Farming Systems. Remote Sens. 2018, 10, 729. [Google Scholar] [CrossRef]
- Mishra, V.N.; Prasad, R.; Rai, P.K.; Vishwakarma, A.K.; Arora, A. Performance Evaluation of Textural Features in Improving Land Use/Land Cover Classification Accuracy of Heterogeneous Landscape Using Multi-Sensor Remote Sensing Data. Earth Sci. Inform. 2019, 12, 71–86. [Google Scholar] [CrossRef]
- Sun, C.; Bian, Y.; Zhou, T.; Pan, J. Using of Multi-Source and Multi-Temporal Remote Sensing Data Improves Crop-Type Mapping in the Subtropical Agriculture Region. Sensors 2019, 19, 2401. [Google Scholar] [CrossRef] [PubMed]
- Tavares, P.A.; Beltrão, N.E.; Guimarães, U.S.; Teodoro, A.C. Integration of Sentinel-1 and Sentinel-2 for Classification and LULC Mapping in the Urban Area of Belém, Eastern Brazilian Amazon. Sensors 2019, 19, 1140. [Google Scholar] [CrossRef] [PubMed]
- Clevers, J.G.P.W.; Gitelson, A.A. Remote Estimation of Crop and Grass Chlorophyll and Nitrogen Content Using Red-Edge Bands on Sentinel-2 and -3. Int. J. Appl. Earth Obs. Geoinf. 2013, 23, 344–351. [Google Scholar] [CrossRef]
- Hall-Beyer, M. Practical Guidelines for Choosing GLCM Textures to Use in Landscape Classification Tasks over a Range of Moderate Spatial Scales. Int. J. Remote Sens. 2017, 38, 1312–1338. [Google Scholar] [CrossRef]
- Haralick, R.; Shanmugam, K.; Dinstein, I. Textural Features for Image Classification. IEEE Trans. Syst. Man Cybern. 1973, 3, 610–621. [Google Scholar] [CrossRef]
- Huete, A.R.; Liu, H.Q.; Batchily, K.; Van Leeuwen, W. A Comparison of Vegetation Indices over a Global Set of TM Images for EOS-MODIS. Remote Sens. Environ. 1997, 59, 440–451. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Kaufman, Y.J.; Merzlyak, M.N. Use of a Green Channel in Remote Sensing of Global Vegetation from EOS-MODIS. Remote Sens. Environ. 1996, 58, 289–298. [Google Scholar] [CrossRef]
- Gao, B.-C. NDWI—A Normalized Difference Water Index for Remote Sensing of Vegetation Liquid Water from Space. Remote Sens. Environ. 1996, 58, 257–266. [Google Scholar] [CrossRef]
- Barnes, E.M.; Clarke, T.; Richards, S.; Colaizzi, P.D.; Haberland, J.; Kostrzewski, M.; Waller, P.; Choi, C.; Riley, E.; Thompson, T. Coincident Detection of Crop Water Stress, Nitrogen Status and Canopy Density Using Ground Based Multispectral Data. In Proceedings of the Fifth International Conference on Precision Agriculture, Bloomington, MN, USA, 16–19 July 2000. [Google Scholar]
- Boiarskii, B.; Hasegawa, H. Comparison of NDVI and NDRE Indices to Detect Differences in Vegetation and Chlorophyll Content. J. Mech. Contin. Math. Sci. 2019, 4, 20–29. [Google Scholar] [CrossRef]
- Rouse, J.W., Jr.; Haas, R.H.; Deering, D.; Schell, J.; Harlan, J.C. Monitoring the Vernal Advancement and Retrogradation (Green Wave Effect) of Natural Vegetation; NASA Goddard Space Flight Center: Greenbelt, MD, USA, 1974; p. 371.
- Merzlyak, M.N.; Gitelson, A.A.; Chivkunova, O.B.; Rakitin, V.Y. Non-Destructive Optical Detection of Pigment Changes during Leaf Senescence and Fruit Ripening. Physiol. Plant. 1999, 106, 135–141. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Gritz, Y.; Merzlyak, M.N. Relationships between Leaf Chlorophyll Content and Spectral Reflectance and Algorithms for Non-Destructive Chlorophyll Assessment in Higher Plant Leaves. J. Plant Physiol. 2003, 160, 271–282. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Merzlyak, M.N. Spectral Reflectance Changes Associated with Autumn Senescence of Aesculus hippocastanum L. and Acer platanoides L. Leaves. Spectral Features and Relation to Chlorophyll Estimation. J. Plant Physiol. 1994, 143, 286–292. [Google Scholar] [CrossRef]
- Huete, A.R. A Soil-Adjusted Vegetation Index (SAVI). Remote Sens. Environ. 1988, 25, 295–309. [Google Scholar] [CrossRef]
- Haboudane, D.; Tremblay, N.; Miller, J. Remote Estimation of Crop Chlorophyll Content Using Spectral Indices Derived from Hyperspectral Data. IEEE Trans. Geosci. Remote Sens. 2008, 46, 423–437. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Kaufman, Y.; Stark, R.; Rundquist, D. Novel Algorithms for Remote Estimation of Vegetation Fraction. Remote Sens. Environ. 2002, 80, 76–87. [Google Scholar] [CrossRef]
- Conners, R.W.; Trivedi, M.M.; Harlow, C.A. Segmentation of a High-Resolution Urban Scene Using Texture Operators. Comput. Vis. Graph. Image Process. 1984, 25, 273–310. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-Learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar] [CrossRef]
- Google Colaboratory Team. Colaboratory. Available online: https://workspace.google.com/marketplace/app/colaboratory/1014160490159 (accessed on 17 September 2023).
- Müllner, D. Modern Hierarchical, Agglomerative Clustering Algorithms. arXiv 2011, arXiv:1109.2378. [Google Scholar] [CrossRef]
- Ward, J.H., Jr. Hierarchical Grouping to Optimize an Objective Function. J. Am. Stat. Assoc. 1963, 58, 236–244. [Google Scholar] [CrossRef]
- Cunningham, P.; Kathirgamanathan, B.; Delany, S.J. Feature Selection Tutorial with Python Examples. arXiv 2021, arXiv:2106.06437. [Google Scholar] [CrossRef]
- Li, J.; Cheng, K.; Wang, S.; Morstatter, F.; Trevino, R.; Tang, J.; Liu, H. Feature Selection: A Data Perspective. ACM Comput. Surv. 2017, 50, 1–45. [Google Scholar] [CrossRef]
- Gregorutti, B.; Michel, B.; Saint-Pierre, P. Correlation and Variable Importance in Random Forests. Stat. Comput. 2017, 27, 659–678. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Strobl, C.; Boulesteix, A.L.; Kneib, T.; Augustin, T.; Zeileis, A. Conditional Variable Importance for Random Forests. BMC Bioinform. 2008, 9, 307. [Google Scholar] [CrossRef]
- Gómez-Ramírez, J.; Ávila-Villanueva, M.; Fernández-Blázquez, M.Á. Selecting the Most Important Self-Assessed Features for Predicting Conversion to Mild Cognitive Impairment with Random Forest and Permutation-Based Methods. Sci. Rep. 2020, 10, 20630. [Google Scholar] [CrossRef]
- Belgiu, M.; Drăguţ, L. Random Forest in Remote Sensing: A Review of Applications and Future Directions. ISPRS J. Photogramm. Remote Sens. 2016, 114, 24–31. [Google Scholar] [CrossRef]
- Deng, C.; Wu, C. The Use of Single-Date MODIS Imagery for Estimating Large-Scale Urban Impervious Surface Fraction with Spectral Mixture Analysis and Machine Learning Techniques. ISPRS J. Photogramm. Remote Sens. 2013, 86, 100–110. [Google Scholar] [CrossRef]
- Du, P.; Samat, A.; Waske, B.; Liu, S.; Li, Z. Random Forest and Rotation Forest for Fully Polarized SAR Image Classification Using Polarimetric and Spatial Features. ISPRS J. Photogramm. Remote Sens. 2015, 105, 38–53. [Google Scholar] [CrossRef]
- Ghimire, B.; Rogan, J.; Galiano, V.R.; Panday, P.; Neeti, N. An Evaluation of Bagging, Boosting, and Random Forests for Land-Cover Classification in Cape Cod, Massachusetts, USA. GISci. Remote Sens. 2012, 49, 623–643. [Google Scholar] [CrossRef]
- Pelletier, C.; Valero, S.; Inglada, J.; Champion, N.; Dedieu, G. Assessing the Robustness of Random Forests to Map Land Cover with High Resolution Satellite Image Time Series over Large Areas. Remote Sens. Environ. 2016, 187, 156–168. [Google Scholar] [CrossRef]
- Condro, A.A.; Setiawan, Y.; Prasetyo, L.B.; Pramulya, R.; Siahaan, L. Retrieving the National Main Commodity Maps in Indonesia Based on High-Resolution Remotely Sensed Data Using Cloud Computing Platform. Land 2020, 9, 377. [Google Scholar] [CrossRef]
- Chan, J.C.-W.; Beckers, P.; Spanhove, T.; Borre, J.V. An Evaluation of Ensemble Classifiers for Mapping Natura 2000 Heathland in Belgium Using Spaceborne Angular Hyperspectral (CHRIS/Proba) Imagery. Int. J. Appl. Earth Obs. Geoinf. 2012, 18, 13–22. [Google Scholar] [CrossRef]
- Erinjery, J.J.; Singh, M.; Kent, R. Mapping and Assessment of Vegetation Types in the Tropical Rainforests of the Western Ghats Using Multispectral Sentinel-2 and SAR Sentinel-1 Satellite Imagery. Remote Sens. Environ. 2018, 216, 345–354. [Google Scholar] [CrossRef]
- Lawrence, R.L.; Moran, C.J. The AmericaView Classification Methods Accuracy Comparison Project: A Rigorous Approach for Model Selection. Remote Sens. Environ. 2015, 170, 115–120. [Google Scholar] [CrossRef]
- Pelletier, C.; Webb, G.I.; Petitjean, F. Temporal Convolutional Neural Network for the Classification of Satellite Image Time Series. Remote Sens. 2019, 11, 523. [Google Scholar] [CrossRef]
- Vaglio Laurin, G.; Liesenberg, V.; Chen, Q.; Guerriero, L.; Del Frate, F.; Bartolini, A.; Coomes, D.; Wilebore, B.; Lindsell, J.; Valentini, R. Optical and SAR Sensor Synergies for Forest and Land Cover Mapping in a Tropical Site in West Africa. Int. J. Appl. Earth Obs. Geoinf. 2013, 21, 7–16. [Google Scholar] [CrossRef]
- Congalton, R.G. A Review of Assessing the Accuracy of Classifications of Remotely Sensed Data. Remote Sens. Environ. 1991, 37, 35–46. [Google Scholar] [CrossRef]
- Pontius, R.G.; Millones, M. Death to Kappa: Birth of Quantity Disagreement and Allocation Disagreement for Accuracy Assessment. Int. J. Remote Sens. 2011, 32, 4407–4429. [Google Scholar] [CrossRef]
- Stehman, S.V. Selecting and Interpreting Measures of Thematic Classification Accuracy. Remote Sens. Environ. 1997, 62, 77–89. [Google Scholar] [CrossRef]
- Inglada, J.; Arias, M.; Tardy, B.; Hagolle, O.; Valero, S.; Morin, D.; Dedieu, G.; Sepulcre, G.; Bontemps, S.; Defourny, P.; et al. Assessment of an Operational System for Crop Type Map Production Using High Temporal and Spatial Resolution Satellite Optical Imagery. Remote Sens. 2015, 7, 12356–12379. [Google Scholar] [CrossRef]
- Inglada, J.; Vincent, A.; Arias, M.; Tardy, B.; Morin, D.; Rodes, I. Operational High Resolution Land Cover Map Production at the Country Scale Using Satellite Image Time Series. Remote Sens. 2017, 9, 95. [Google Scholar] [CrossRef]
- Lyons, M.B.; Keith, D.A.; Phinn, S.R.; Mason, T.J.; Elith, J. A Comparison of Resampling Methods for Remote Sensing Classification and Accuracy Assessment. Remote Sens. Environ. 2018, 208, 145–153. [Google Scholar] [CrossRef]
- Google Earth. Google Earth Versions. Available online: https://www.google.com/intl/en/earth/versions/ (accessed on 17 September 2023).
- QGIS.org. QGIS Geographic Information System. Open Source Geospatial Foundation Project. Available online: http://qgis.org (accessed on 17 September 2023).
- Hermosilla, T.; Wulder, M.A.; White, J.C.; Coops, N.C. Land Cover Classification in an Era of Big and Open Data: Optimizing Localized Implementation and Training Data Selection to Improve Mapping Outcomes. Remote Sens. Environ. 2022, 268, 112780. [Google Scholar] [CrossRef]
- Pelletier, C.; Valero, S.; Inglada, J.; Champion, N.; Marais Sicre, C.; Dedieu, G. Effect of Training Class Label Noise on Classification Performances for Land Cover Mapping with Satellite Image Time Series. Remote Sens. 2017, 9, 173. [Google Scholar] [CrossRef]
- Qin, R.; Liu, T. A Review of Landcover Classification with Very-High Resolution Remotely Sensed Optical Images—Analysis Unit, Model Scalability and Transferability. Remote Sens. 2022, 14, 646. [Google Scholar] [CrossRef]
- Anyimah, F.O.; Osei, E.M., Jr.; Nyamekye, C. Detection of Stress Areas in Cocoa Farms Using GIS and Remote Sensing: A Case Study of Offinso Municipal & Offinso North District, Ghana. Environ. Chall. 2021, 4, 100087. [Google Scholar] [CrossRef]
- Millard, K.; Richardson, M. On the Importance of Training Data Sample Selection in Random Forest Image Classification: A Case Study in Peatland Ecosystem Mapping. Remote Sens. 2015, 7, 8489–8515. [Google Scholar] [CrossRef]
- Chen, D.; Stow, D.A.; Gong, P. Examining the Effect of Spatial Resolution and Texture Window Size on Classification Accuracy: An Urban Environment Case. Int. J. Remote Sens. 2004, 25, 2177–2192. [Google Scholar] [CrossRef]
- Hall-Beyer, M. GLCM Texture: A Tutorial, 3rd ed.; Department of Geography, University of Calgary: Calgary, AB, Canada, 2017. [Google Scholar]
- Roberts, D.; Mueller, N.; Mcintyre, A. High-Dimensional Pixel Composites from Earth Observation Time Series. IEEE Trans. Geosci. Remote Sens. 2017, 55, 6254–6264. [Google Scholar] [CrossRef]
- Bey, A.; Jetimane, J.; Lisboa, S.N.; Ribeiro, N.; Sitoe, A.; Meyfroidt, P. Mapping Smallholder and Large-Scale Cropland Dynamics with a Flexible Classification System and Pixel-Based Composites in an Emerging Frontier of Mozambique. Remote Sens. Environ. 2020, 239, 111611. [Google Scholar] [CrossRef]
- Li, Q.; Qiu, C.; Ma, L.; Schmitt, M.; Zhu, X.X. Mapping the Land Cover of Africa at 10 m Resolution from Multi-Source Remote Sensing Data with Google Earth Engine. Remote Sens. 2020, 12, 602. [Google Scholar] [CrossRef]
- Scott, G.J.; England, M.; Starms, W.A.; Marcum, R.A.; Davis, C.H. Training Deep Convolutional Neural Networks for Land-Cover Classification of High-Resolution Imagery. IEEE Geosci. Remote Sens. Lett. 2017, 14, 549–553. [Google Scholar] [CrossRef]
- Tuia, D.; Volpi, M.; Copa, L.; Kanevski, M.F.; Muñoz-Marí, J. A Survey of Active Learning Algorithms for Supervised Remote Sensing Image Classification. IEEE J. Sel. Top. Signal Process. 2011, 5, 606–617. [Google Scholar] [CrossRef]
- Turkoglu, M.O.; D’Aronco, S.; Perich, G.; Liebisch, F.; Streit, C.; Schindler, K.; Wegner, J.D. Crop Mapping from Image Time Series: Deep Learning with Multi-Scale Label Hierarchies. Remote Sens. Environ. 2021, 264, 112603. [Google Scholar] [CrossRef]
- Crawford, M.M.; Tuia, D.; Yang, H.L. Active Learning: Any Value for Classification of Remotely Sensed Data? Proc. IEEE 2013, 101, 593–608. [Google Scholar] [CrossRef]
- Critchley, M.; Sassen, M.; Umunay, P. Mapping Opportunities for Cocoa Agroforestry in Côte d’Ivoire: Assessing its Potential to Contribute to National Forest Cover Restoration Targets and Ecosystem Services Co-Benefits; UNEP World Conservation Monitoring Centre: Cambridge, UK, 2021. [Google Scholar]
- Mighty Earth. Sweet Nothings: How the Chocolate Industry Has Failed to Honor Promises to End Deforestation for Cocoa in Côte d’Ivoire and Ghana; Mighty Earth: Washington, DC, USA, 2022. [Google Scholar]
- Copernicus. Copernicus HotSpot Land Cover Change Explorer. Available online: https://land.copernicus.eu/global/hsm (accessed on 27 September 2023).







| Data | Reference Dataset | Cocoa Pixels | Cocoa Polygons | Non-Cocoa Pixels |
|---|---|---|---|---|
| Training | Raw | 9427 | 68 | 9322 |
| Refined | 4437 | 4392 | ||
| Validation | Raw | 405 | 14 | 405 |
| Refined | 186 | 186 | ||
| Test | Refined | 723 | 11 | 719 |
| Reference Dataset | Input Data | OA (%) | Cocoa | ||
|---|---|---|---|---|---|
| UA (%) | PA (%) | F1 (%) | |||
| Refined | 4 features 1 | 80.6 ± 1.6 | 82.4 ± 1.2 | 77.9 ± 2.8 | 80.1 ± 1.9 |
| 4 features 1 + 12 Sentinel-2 bands | 85.1 ± 2.0 | 87.5 ± 0.8 | 81.9 ± 3.9 | 84.6 ± 2.4 | |
| 25 features 2 | 82.3 ± 1.8 | 87.0 ± 0.8 | 75.9 ± 3.6 | 81.1 ± 2.3 | |
| 25 features 2 + 12 Sentinel-2 bands | 84.7 ± 1.9 | 88.9 ± 0.9 | 79.3 ± 3.5 | 83.8 ± 2.3 | |
| 12 Sentinel-2 bands | 84.3 ± 2.4 | 87.8 ± 1.1 | 79.9 ± 4.6 | 83.6 ± 2.9 | |
| Raw | 4 features 3 | 82.1 ± 1.8 | 80.4 ± 1.4 | 84.7 ± 3.4 | 82.4 ± 2.2 |
| 4 features 3 + 12 Sentinel-2 bands | 83.4 ± 2.2 | 85.0 ± 1.1 | 81.2 ± 4.1 | 83.0 ± 2.5 | |
| 25 features 4 | 80.7 ± 3.2 | 85.5 ± 1.4 | 73.9 ± 6.2 | 79.2 ± 4.1 | |
| 25 features 4 + 12 Sentinel-2 bands | 83.9 ± 2.6 | 87.7 ± 1.2 | 78.8 ± 5.0 | 83.0 ± 3.2 | |
| 12 Sentinel-2 bands | 83.7 ± 2.3 | 85.8 ± 1.2 | 80.9 ± 4.2 | 83.2 ± 2.7 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moraiti, N.; Mullissa, A.; Rahn, E.; Sassen, M.; Reiche, J. Critical Assessment of Cocoa Classification with Limited Reference Data: A Study in Côte d’Ivoire and Ghana Using Sentinel-2 and Random Forest Model. Remote Sens. 2024, 16, 598. https://0-doi-org.brum.beds.ac.uk/10.3390/rs16030598
Moraiti N, Mullissa A, Rahn E, Sassen M, Reiche J. Critical Assessment of Cocoa Classification with Limited Reference Data: A Study in Côte d’Ivoire and Ghana Using Sentinel-2 and Random Forest Model. Remote Sensing. 2024; 16(3):598. https://0-doi-org.brum.beds.ac.uk/10.3390/rs16030598
Chicago/Turabian StyleMoraiti, Nikoletta, Adugna Mullissa, Eric Rahn, Marieke Sassen, and Johannes Reiche. 2024. "Critical Assessment of Cocoa Classification with Limited Reference Data: A Study in Côte d’Ivoire and Ghana Using Sentinel-2 and Random Forest Model" Remote Sensing 16, no. 3: 598. https://0-doi-org.brum.beds.ac.uk/10.3390/rs16030598