The Interplay between Nutrition, Innate Immunity, and the Commensal Microbiota in Adaptive Intestinal Morphogenesis
Abstract
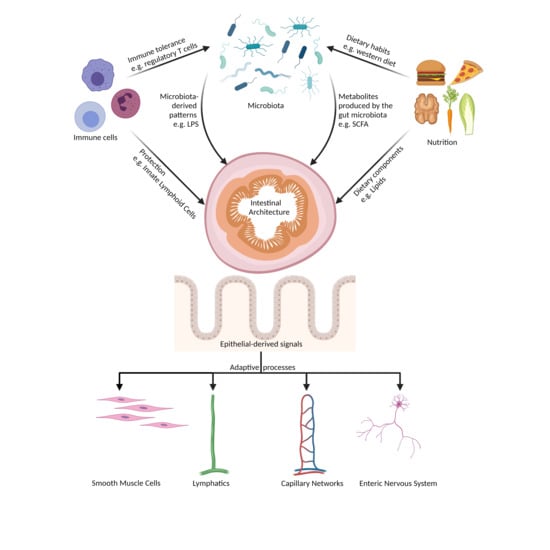
:1. Introduction
2. Development of Intestinal Tissues
2.1. General Aspects and Early Gut Development
2.2. Development of the Intestinal Epithelium and Villus Formation
2.3. Intestinal Vasculogenesis
2.4. Development of the Enteric Nervous System
3. Intestinal Epithelial Homeostasis and Renewal
3.1. Epithelial Lineage Commitment in the Self-Renewing Crypt-Villus Unit
| Cell Type (Ref.) | Localization | Histomorphology | Key Functions |
|---|---|---|---|
| Enterocyte [101] | Small intestine, large intestine Along villus | Columnar polarized cell with basal oval nucleus Apical brush border (Microvilli) | Absorption of nutrients, water Gut barrier Secretion of antimicrobial peptides Interaction with innate immune system |
| Goblet cell [100] | Small intestine, Colon Along villus | Columnar cell, apical part enlarged Small triangular nucleus basal Cytoplasm with secretory granules (mucins) | Secretion of mucins and other glycoproteins Nonspecific endocytosis of antigens Interaction with innate immune system |
| Enteroendocrine cell [102] | Small intestine, Colon Along villus | Shape varies with cell subtype, based on secreted hormones Long basal processes to interface with neurons or adjacent intestinal epithelial cells | Secrete hormones (e.g., serotonin) Detect gut microbes and microbial metabolites Interaction with innate immune cells |
| Tuft cell [103,104] | Small intestine, Colon Along villus | Cylindrical cell body, narrows on apical and basal end Highly organized brush border Lateral membrane projections to adjacent enterocytes | Innate immune responses to helminth infection Contribute to epithelial regeneration |
| Microfold cell [105] | Small intestine Above gut associated lymphatic tissue (GALT) | Columnar cell No apical brush border Basal interaction with lymphocytes/dendritic cells | Capture luminal antigen and present it to immune cells Inflammation can induce development of microfold cells |
| Paneth cell [96] | Small intestine Crypts | Large eosinophilic secretory granules | Secrete antimicrobial peptides Regulating stem cell niche |
3.2. The Functional Role of the Intestinal Mucus Layer
4. Nutrition, Microbiota, and Innate Immune Signaling Adapts Gut Morphology and Cell Homeostasis
4.1. Effects on Epithelial Lineage Commitment and Renewal
4.1.1. Impact of Nutrition
4.1.2. Impact of Innate Immune Functions
4.1.3. Impact of the Gut Microbiome
4.2. Effects on the Intestinal Microvasculature
4.2.1. Impact of Nutrition
4.2.2. Impact of Innate Immune Functions
4.2.3. Impact of the Gut Microbiome
4.3. Effects on the Enteric Nervous System
4.3.1. Impact of Nutrition
4.3.2. Impact of Innate Immune Functions
4.3.3. Impact of the Gut Microbiome
4.4. Effects on Intestinal Smooth Muscle Cell Layers
4.4.1. Impact of Nutrition
4.4.2. Impact of Innate Immune Functions
4.4.3. Impact of the Gut Microbiome
5. Challenges and Limitations
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kiefer, J.C. Molecular mechanisms of early gut organogenesis: A primer on development of the digestive tract. Dev. Dyn. 2003, 228, 287–291. [Google Scholar] [CrossRef]
- Noah, T.K.; Donahue, B.; Shroyer, N.F. Intestinal development and differentiation. Exp. Cell Res. 2011, 317, 2702–2710. [Google Scholar] [CrossRef] [Green Version]
- Ziegler, A.L.; Pridgen, T.A.; Blikslager, A.T. Environmental stressors affect intestinal permeability and repair responses in a pig intestinal ischemia model. Tissue Barriers 2020, 8, 1832421. [Google Scholar] [CrossRef]
- Gribble, F.M.; Reimann, F. Metabolic messengers: Glucagon-like peptide 1. Nat. Metab. 2021, 3, 142–148. [Google Scholar] [CrossRef]
- Collado, M.C.; Rautava, S.; Aakko, J.; Isolauri, E.; Salminen, S. Human gut colonisation may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid. Sci. Rep. 2016, 6, 23129. [Google Scholar] [CrossRef] [Green Version]
- De Goffau, M.C.; Lager, S.; Sovio, U.; Gaccioli, F.; Cook, E.; Peacock, S.J.; Parkhill, J.; Charnock-Jones, D.S.; Smith, G.C.S. Human placenta has no microbiome but can contain potential pathogens. Nature 2019, 572, 329–334. [Google Scholar] [CrossRef] [PubMed]
- Blaser, M.J.; Devkota, S.; McCoy, K.D.; Relman, D.A.; Yassour, M.; Young, V.B. Lessons learned from the prenatal microbiome controversy. Microbiome 2021, 9, 8. [Google Scholar] [CrossRef]
- Reinhardt, C.; Reigstad, C.S.; Bäckhed, F. Intestinal microbiota during infancy and ist implications for obesity. J. Pediatr. Gastroenterol. Nutr. 2007, 48, 249–256. [Google Scholar] [CrossRef] [Green Version]
- Roswall, J.; Olsson, L.M.; Kovatcheva-Datchary, P.; Nilsson, S.; Tremaroli, V.; Simon, M.C.; Kiilerich, P.; Krämer, M.; Uhlén, M.; Gummesson, A.; et al. Developmental trajectory of the healty human gut microbiota during the first 5 years of life. Cell Host. Microbe 2021, 29, 765–776.e3. [Google Scholar] [CrossRef]
- Bäckhed, F.; Ley, R.E.; Sonnenburg, J.L.; Peterson, D.A.; Gordon, J.I. Host-bacterial mutualism in the human intestine. Science 2005, 307, 1915–1920. [Google Scholar] [CrossRef] [Green Version]
- Radaic, A.; Kapila, Y.L. The oralome and its dysbiosis: New insights into oral microbiome-host interactions. Comput. Struct. Biotechnol. J. 2021, 19, 1335–1360. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Dale, B.A. Activation of protective responses in oral epithelial cells by Fusobacterium nucleatum and human beta-defensin-2. J. Med. Microbiol. 2007, 56 Pt 7, 976–987. [Google Scholar] [CrossRef] [Green Version]
- Ganal-Vonarburg, S.C.; Duerr, C.U. The interaction of intestinal microbiota and innate lymphoid cells in health and disease throughout life. Immunology 2020, 159, 39–51. [Google Scholar] [CrossRef] [PubMed]
- Fåk, F.; Ahrné, S.; Molin, G.; Jeppsson, B.; Weström, B. Microbial manipulation of the rat dam changes bacterial colonization and alters properties of the gut in her offspring. Am. J. Physiol. Gastrointest. Liver. Physiol. 2008, 294, G148–G154. [Google Scholar] [CrossRef] [PubMed]
- Alsharairi, N.A. The infant gut microbiota and risk of asthma: The effect of maternal nutrition during pregnancy and lactation. Microorganisms 2020, 8, 1119. [Google Scholar] [CrossRef] [PubMed]
- Gomes-Neto, J.C.; Kittana, H.; Mantz, S.; Segura Munoz, R.R.; Schmaltz, R.J.; Bindels, L.B.; Clarke, J.; Hostetter, J.M.; Benson, A.K.; Walter, J.; et al. A gut pathobiont synergizes with the microbiota to instigate inflammatory disease marked by immunoreactivity against other symbionts but not itself. Sci. Rep. 2017, 7, 17707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, R.K.; Chang, H.-W.; Yan, D.; Lee, K.M.; Ucmak, D.; Wong, K.; Abrouk, M.; Farahnik, B.; Nakamura, M.; Zhu, T.H.; et al. Influence of diet on the gut microbiome and implications for human health. J. Transl. Med. 2017, 15, 73. [Google Scholar] [CrossRef] [Green Version]
- Venter, C.; Eyerich, S.; Sarin, T.; Klatt, K.C. Nutrition and the immune system: A complicated tango. Nutrients 2020, 12, 818. [Google Scholar] [CrossRef] [Green Version]
- Van der Flier, L.G.; Clevers, H. Stem cells, self-renewal, and differentiation in the intestinal epithelium. Annu. Rev. Physiol. 2009, 71, 241–260. [Google Scholar] [CrossRef]
- Probst, H.C.; Muth, S.; Schild, H. Regulation of the tolerogenic function of steady-state DCs. Eur. J. Immunol. 2014, 44, 927–933. [Google Scholar] [CrossRef]
- Schaupp, L.; Muth, S.; Rogell, L.; Kofoed-Branzk, M.; Melchior, F.; Lienenklaus, S.; Ganal-Vonarburg, S.C.; Klein, M.; Guendel, F.; Hain, T.; et al. Microbiota-Induced Type I Interferons Instruct a Poised Basal State of Dendritic Cells. Cell 2020, 181, 1080–1096.e19. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, M.; Hara, K.; Benno, Y. The influence of the immunostimulation by bacterial cell components derived from altered large intestinal microbiota on probiotic anti-inflammatory benefits. FEMS Immunol. Med. Microbiol. 2007, 49, 387–390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carriche, G.M.; Almeida, L.; Stüve, P.; Velasquez, L.; Dhillon-LaBrooy, A.; Roy, U.; Lindenberg, M.; Strowig, T.; Plaza-Sirvent, C.; Schmitz, I.; et al. Regulating T-cell differentiation through the polyamine spermidine. J. Allergy Clin. Immunol. 2021, 147, 335–348.e11. [Google Scholar] [CrossRef] [PubMed]
- Johnson, E.L.; Heaver, S.L.; Waters, J.L.; Kim, B.I.; Bretin, A.; Goodman, A.L.; Gewirtz, A.T.; Worgall, T.S.; Ley, R.E. Sphingolipids produced by gut bacteria enter host metabolic pathways impacting ceramide levels. Nat. Commun. 2020, 11, 2471. [Google Scholar] [CrossRef]
- Koh, A.; Bäckhed, F. From association to causality: The role of the gut microbiota and its functional products on host metabolism. Mol. Cell 2020, 78, 584–596. [Google Scholar] [CrossRef]
- Arpaia, N.; Campbell, C.; Fan, X.; Dikiy, S.; van der Veeken, J.; deRoos, P.; Liu, H.; Cross, J.R.; Pfeffer, K.; Coffer, P.J.; et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 2013, 504, 451–455. [Google Scholar] [CrossRef]
- Schroeder, B.O.; Bäckhed, F. Signals from the gut microbiota to distant organs in physiology and disease. Nat. Med. 2016, 22, 1079–1089. [Google Scholar] [CrossRef]
- Jäckel, S.; Kiouptsi, K.; Lillich, M.; Hendrikx, T.; Khandagale, A.; Kollar, B.; Hörmann, N.; Reiss, C.; Subramaniam, S.; Wilms, E.; et al. Gut microbiota regulate hepatic von Willebrand factor synthesis and arterial thrombus formation via Toll-like receptor-2. Blood 2017, 130, 542–553. [Google Scholar] [CrossRef] [Green Version]
- Nigro, G.; Rossi, R.; Commere, P.H.; Jay, P.; Sansonetti, P.J. The cytosolic bacterial peptidoglycan sensor Nod2 affords stem cell protection and links microbes to gut epithelial regeneration. Cell Host Microbe 2014, 15, 792–798. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hörmann, N.; Brandão, I.; Jäckel, S.; Ens, N.; Lillich, M.; Walter, U.; Reinhardt, C. Gut microbial colonization orchestrates TLR2 expression, signaling and epithelial proliferation in the small intestinal mucosa. PLoS ONE 2014, 9, e113080. [Google Scholar] [CrossRef] [Green Version]
- Stappenbeck, T.S.; Hooper, L.V.; Gordon, J.I. Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc. Natl. Acad. Sci. USA 2002, 99, 15451–15455. [Google Scholar] [CrossRef] [Green Version]
- Reinhardt, C.; Bergentall, M.; Greiner, T.U.; Schaffner, F.; Ostergren-Lundén, G.; Petersen, L.C.; Ruf, W.; Bäckhed, F. Tissue factor and PAR1 promote microbiota-induced intestinal vascular remodelling. Nature 2012, 483, 627–631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suh, S.H.; Choe, K.; Hong, S.P.; Jeong, S.H.; Mäkinen, T.; Kim, K.S.; Alitalo, K.; Surh, C.D.; Koh, G.Y.; Song, J.H. Gut microbiota regulates lacteal integrity by inducing VEGF-C in intestinal villus macrophages. EMBO Rep. 2019, 20, e46927. [Google Scholar] [CrossRef] [PubMed]
- Anitha, M.; Vijay-Kumar, M.; Sitaraman, S.V.; Gewirtz, A.T.; Srinivasan, S. Gut microbial products regulate murine gastrointestinal motility via Toll-like receptor 4 signaling. Gastroenterology 2012, 143, 1006–1016.e4. [Google Scholar] [CrossRef] [Green Version]
- Martini, E.; Krug, S.M.; Siegmund, B.; Neurath, M.F.; Becker, C. Mend your fences: The epithelial barrier and its relationship with mucosal immunity in inflammatory bowel disease. Cell. Mol. Gastroenterol. Hepatol. 2017, 4, 33–46. [Google Scholar] [CrossRef] [Green Version]
- Arvidsson, C.; Hallén, A.; Bäckhed, F. Generating and analyzing germ-free mice. Curr. Protoc. Mouse Biol. 2012, 2, 307–316. [Google Scholar] [CrossRef]
- Kaufman, M.H. The Atlas of Mouse Development; Academic Press: London, UK, 1995; plate 15a. [Google Scholar]
- Spence, J.R.; Lauf, R.; Shroyer, N.F. Vertebrate intestinal endoderm development. Dev. Dyn. 2011, 240, 501–520. [Google Scholar] [CrossRef] [Green Version]
- Van den Brink, G.R. Hedgehog signaling in development and homeostasis of the gastrointestinal tract. Physiol. Rev. 2007, 87, 1343–1375. [Google Scholar] [CrossRef]
- Shook, D.; Keller, R. Mechanisms, mechanics and function of epithelial-mesenchymal transitions in early development. Mech. Dev. 2003, 120, 1351–1383. [Google Scholar] [CrossRef] [PubMed]
- McLin, V.A.; Rankin, S.A.; Zorn, A.M. Repression of Wnt/beta-catenin signaling in the anterior endoderm is essential for liver and pancreas development. Development 2007, 134, 2207–2217. [Google Scholar] [CrossRef] [Green Version]
- Sherwood, R.I.; Chen, T.Y.; Melton, D.A. Transcriptional dynamics of endodermal organ formation. Dev. Dyn. 2009, 238, 29–42. [Google Scholar] [CrossRef] [Green Version]
- Grainger, S.; Savory, J.G.; Lohnes, D. Cdx2 regulates patterning of the intestinal epithelium. Dev. Biol. 2010, 339, 155–165. [Google Scholar] [CrossRef] [Green Version]
- Ramalho-Santos, M.; Melton, D.A.; McMahon, A.P. Hedgehog signals regulate multiple aspects of gastrointestinal development. Development 2000, 127, 2763–2772. [Google Scholar] [CrossRef]
- Grosse, A.S.; Pressprich, M.F.; Curley, L.B.; Hamilton, K.L.; Margolis, B.; Hildebrand, J.D.; Gumucio, D.L. Cell dynamics in fetal intestinal epithelium: Implications for intestinal growth and morphogenesis. Development 2011, 138, 4423–4432. [Google Scholar] [CrossRef] [Green Version]
- Lepourcelet, M.; Tou, L.; Cai, L.; Sawada, J.; Lazar, A.J.; Glickman, J.N.; Williamson, J.A.; Everett, A.D.; Redston, M.; Fox, E.A.; et al. Insights into developmental mechanisms and cancers in the mammalian intestine derived from serial analysis of gene expression and study of the hepatoma-derived growth factor (HDGF). Development 2005, 132, 415–427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cervantes, S.; Yamaguchi, T.P.; Hebrok, M. Wnt5a is essential for intestinal elongation in mice. Dev. Biol. 2009, 326, 285–294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qian, D.; Jones, C.; Rzadzinska, A.; Mark, S.; Zhang, X.; Steel, K.P.; Dai, X.; Chen, P. Wnt5a functions in planar cell polarity regulation in mice. Dev. Biol. 2007, 306, 121–133. [Google Scholar] [CrossRef] [Green Version]
- Mao, J.; Kim, B.M.; Rajurkar, M.; Shivdasani, R.A.; McMahon, A.P. Hedgehog signaling controls mesenchymal growth in the developing mammalian digestive tract. Development 2010, 137, 1721–1729. [Google Scholar] [CrossRef] [Green Version]
- Helander, H.F. Morphological studies on the development of the rat colonic mucosa. Acta Anat. 1973, 85, 155–176. [Google Scholar] [CrossRef] [PubMed]
- Walton, K.D.; Mishkind, D.; Riddle, M.R.; Tabin, C.J.; Gumucio, D.L. Blueprint for an intestinal villus: Species-specific assembly required. Wiley Interdiscip. Rev. Dev. Biol. 2018, 7, e317. [Google Scholar] [CrossRef]
- Walton, K.D.; Kolterud, A.; Czerwinski, M.J.; Bell, M.J.; Prakash, A.; Kushwaha, J.; Grosse, A.S.; Schnell, S.; Gumucio, D.L. Hedgehog-responsive mesenchymal clusters direct patterning and emergence of intestinal villi. Proc. Natl. Acad. Sci. USA 2012, 109, 15817–15822. [Google Scholar] [CrossRef] [Green Version]
- Karlsson, L.; Lindahl, P.; Heath, J.K.; Betsholtz, C. Abnormal gastrointestinal development in PDGF-A and PDGFR-(alpha) deficient mice implicates a novel mesenchymal structure with putative instructive properties in villus morphogenesis. Development 2000, 127, 3457–3466. [Google Scholar] [CrossRef]
- Madison, B.B.; Braunstein, K.; Kuizon, E.; Portman, K.; Qiao, X.T.; Gumucio, D.L. Epithelial hedgehog signals pattern the intestinal crypt-villus axis. Development 2005, 132, 279–289. [Google Scholar] [CrossRef] [Green Version]
- Calvert, R.; Micheletti, P.A. Selection of a chemically defined medium for culturing fetal mouse small intestine. In Vitro 1981, 17, 331–344. [Google Scholar] [CrossRef] [PubMed]
- Kolterud, A.; Grosse, A.S.; Zacharias, W.J.; Walton, K.D.; Kretovich, K.E.; Madison, B.B.; Waghray, M.; Ferris, J.E.; Hu, C.; Merchant, J.L.; et al. Paracrine Hedgehog signaling in stomach and intestine: New roles for hedgehog in gastrointestinal patterning. Gastroenterology 2009, 137, 618–628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, H.; Cotton, J.L.; Wang, Y.; Rajurkar, M.; Zhu, L.J.; Lewis, B.C.; Mao, J. Specific requirement of Gli transcription factors in Hedgehog-mediated intestinal development. J. Biol. Chem. 2013, 288, 17589–17596. [Google Scholar] [CrossRef] [Green Version]
- Walton, K.D.; Whidden, M.; Kolterud, Å.; Shoffner, S.K.; Czerwinski, M.J.; Kushwaha, J.; Parmar, N.; Chandhrasekhar, D.; Freddo, A.M.; Schnell, S.; et al. Villification in the mouse: Bmp signals control intestinal villus patterning. Development 2016, 143, 427–436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Freddo, A.M.; Shoffner, S.K.; Shao, Y.; Taniguchi, K.; Grosse, A.S.; Guysinger, M.N.; Wang, S.; Rudraraju, S.; Margolis, B.; Garikipati, K.; et al. Coordination of signaling and tissue mechanics during morphogenesis of murine intestinal villi: A role for mitotic cell rounding. Integr. Biol. 2016, 8, 918–928. [Google Scholar] [CrossRef] [PubMed]
- Watson, C.L.; Mahe, M.M.; Múnera, J.; Howell, J.C.; Sundaram, N.; Poling, H.M.; Schweitzer, J.I.; Vallance, J.E.; Mayhew, C.N.; Sun, Y.; et al. An in vivo model of human small intestine using pluripotent stem cells. Nat. Med. 2014, 20, 1310–1314. [Google Scholar] [CrossRef] [Green Version]
- Wilm, B.; Ipenberg, A.; Hastie, N.D.; Burch, J.B.; Bader, D.M. The serosal mesothelium is a major source of smooth muscle cells of the gut vasculature. Development 2005, 132, 5317–5328. [Google Scholar] [CrossRef] [Green Version]
- Hatch, J.; Mukouyama, Y.S. Spatiotemporal mapping of vascularization and innervation in the fetal murine intestine. Dev. Dyn. 2015, 244, 56–68. [Google Scholar] [CrossRef]
- BioRender. Available online: https://app.biorender.com (accessed on 25 June 2021).
- Bäckhed, F.; Crawford, P.A.; O’Donnell, D.; Gordon, J.I. Postnatal lymphatic partitioning from the blood vasculature in the small intestine requires fasting-induced adipose factor. Proc. Natl. Acad. Sci. USA 2007, 104, 606–611. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niesler, B.; Kuerten, S.; Demir, I.E.; Schäfer, K.H. Disorders of the enteric nervous system—a holistic view. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 393–410. [Google Scholar] [CrossRef]
- Sharkey, K.A. Emerging roles for enteric glia in gastrointestinal disorders. J. Clin. Invest. 2015, 125, 918–925. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Endres, K.; Schäfer, K.H. Influence of commensal microbiota on the enteric nervous system and its role in neurodegenerative diseases. J. Innate Immun. 2018, 10, 172–180. [Google Scholar] [CrossRef] [PubMed]
- Young, H.M.; Hearn, C.J.; Newgreen, D.F. Embryology and development of the enteric nervous system. Gut 2000, 47 (Suppl. S4), iv12–iv14, discussion iv26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le Douarin, N.M.; Teillet, M.A. The migration of neural crest cells to the wall of the digestive tract in avian embryo. J. Embryol. Exp. Morphol. 1973, 30, 31–48. [Google Scholar]
- Young, H.M.; Newgreen, D. Enteric neural crest-derived cells: Origin, identification, migration, and differentiation. Anat. Rec. 2001, 262, 1–15. [Google Scholar] [CrossRef]
- Young, H.M.; Hearn, C.J.; Ciampoli, D.; Southwell, B.R.; Brunet, J.F.; Newgreen, D.F. A single rostrocaudal colonization of the rodent intestine by enteric neuron precursors is revealed by the expression of Phox2b, Ret, and p75 and by explants grown under the kidney capsule or in organ culture. Dev. Biol. 1998, 202, 67–84. [Google Scholar] [CrossRef] [Green Version]
- Fu, M.; Lui, V.C.; Sham, M.H.; Cheung, A.N.; Tam, P.K. HOXB5 expression is spatially and temporarily regulated in human embryonic gut during neural crest cell colonization and differentiation of enteric neuroblasts. Dev. Dyn. 2003, 228, 1–10. [Google Scholar] [CrossRef]
- Gershon, M.D. Genes and lineages in the formation of the enteric nervous system. Curr. Opin. Neurobiol. 1997, 7, 101–109. [Google Scholar] [CrossRef]
- Timmermans, M.W.; Scheuermann, D.W. Distributional pattern and targets of GABA-containing neurons in the porcine small and large intestine. Eur. J. Morphol. 1998, 36, 133–142. [Google Scholar] [CrossRef] [PubMed]
- Bergner, A.J.; Stamp, L.A.; Gonsalvez, D.G.; Allison, M.B.; Olson, D.P.; Myers, M.G., Jr.; Anderson, C.R.; Young, H.M. Birthdating of myenteric neuron subtypes in the small intestine of the mouse. J. Comp. Neurol. 2014, 522, 514–527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Erickson, C.S.; Lee, S.J.; Barlow-Anacker, A.J.; Druckenbrod, N.R.; Epstein, M.L.; Gosain, A. Appearance of cholinergic myenteric neurons during enteric nervous system development: Comparison of different ChAT fluorescent mouse reporter lines. Neurogastroenterol. Motil. 2014, 26, 874–884. [Google Scholar] [CrossRef] [Green Version]
- Pham, T.D.; Gershon, M.D.; Rothman, T.P. Time of origin of neurons in the murine enteric nervous system: Sequence in relation to phenotype. J. Comp. Neurol. 1991, 314, 789–798. [Google Scholar] [CrossRef] [PubMed]
- Belkind-Gerson, J.; Carreon-Rodriguez, A.; Benedict, L.A.; Steiger, C.; Pieretti, A.; Nagy, N.; Dietrich, J.; Goldstein, A.M. Nestin-expressing cells in the gut give rise to enteric neurons and glial cells. Neurogastroenterol. Motil. 2013, 25, 61–69.e7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laranjeira, C.; Sandgren, K.; Kessaris, N.; Richardson, W.; Potocnik, A.; Vanden Berghe, P.; Pachnis, V. Glial cells in the mouse enteric nervous system can undergo neurogenesis in response to injury. J. Clin. Investig. 2011, 121, 3412–3424. [Google Scholar] [CrossRef] [Green Version]
- Delalande, J.M.; Natarajan, D.; Vernay, B.; Finlay, M.; Ruhrberg, C.; Thapar, N.; Burns, A.J. Vascularisation is not necessary for gut colonisation by enteric neural crest cells. Dev. Biol. 2014, 385, 220–229. [Google Scholar] [CrossRef] [Green Version]
- Schrenk, S.; Schuster, A.; Klotz, M.; Schleser, F.; Lake, J.; Heuckeroth, R.O.; Kim, Y.J.; Laschke, M.W.; Menger, M.D.; Schäfer, K.H. Vascular and neural stem cells in the gut: Do they need each other? Histochem. Cell Biol. 2015, 143, 397–410. [Google Scholar] [CrossRef]
- Peterson, L.W.; Artis, D. Intestinal epithelial cells: Regulators of barrier function and immune homeostasis. Nat. Rev. Immunol. 2014, 14, 141–153. [Google Scholar] [CrossRef]
- Spadoni, I.; Zagato, E.; Bertocchi, A.; Paolinelli, R.; Hot, E.; Di Sabatino, A.; Caprioli, F.; Bottiglieri, L.; Oldani, A.; Viale, G.; et al. A gut-vascular barrier controls the systemic dissemination of bacteria. Science 2015, 350, 830–834. [Google Scholar] [CrossRef]
- Mowat, A.M.; Agace, W.W. Regional specialization within the intestinal immune system. Nat. Rev. Immunol. 2014, 14, 667–685. [Google Scholar] [CrossRef] [PubMed]
- Clevers, H. The intestinal crypt, a prototype stem cell compartment. Cell 2013, 154, 274–284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bischoff, S.C.; Barbara, G.; Buurman, W.; Ockhuizen, T.; Schulzke, J.D.; Serino, M.; Tilg, H.; Watson, A.; Wells, J.M. Intestinal permeability—A new target for disease prevention and therapy. BMC Gastroenterol. 2014, 14, 189. [Google Scholar] [CrossRef] [Green Version]
- Gu, Y.; Forostyan, T.; Sabbadini, R.; Rosenblatt, J. Epithelial cell extrusion requires the sphingosine-1-phosphate receptor 2 pathway. J. Cell Biol. 2011, 193, 667–676. [Google Scholar] [CrossRef]
- Bjerknes, M.; Cheng, H. Clonal analysis of mouse intestinal epithelial progenitors. Gastroenterology 1999, 116, 7–14. [Google Scholar] [CrossRef]
- Potten, C.S.; Kovacs, L.; Hamilton, E. Continuous labelling studies on mouse skin and intestine. Cell Tissue Kinet. 1974, 7, 271–283. [Google Scholar] [CrossRef]
- Cheng, H.; Leblond, C.P. Origin, differentiation and renewal of the four main epithelial cell types in the mouse small intestine. V. Unitarian Theory of the origin of the four epithelial cell types. Am. J. Anat. 1974, 141, 537–561. [Google Scholar] [CrossRef]
- Barker, N.; van Es, J.H.; Kuipers, J.; Kujala, P.; van den Born, M.; Cozijnsen, M.; Haegebarth, A.; Korving, J.; Begthel, H.; Peters, P.J.; et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 2007, 449, 1003–1007. [Google Scholar] [CrossRef]
- Snippert, H.J.; van der Flier, L.G.; Sato, T.; van Es, J.H.; van den Born, M.; Kroon-Veenboer, C.; Barker, N.; Klein, A.M.; van Rheenen, J.; Simons, B.D.; et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell 2010, 143, 134–144. [Google Scholar] [CrossRef] [Green Version]
- Milano, J.; McKay, J.; Dagenais, C.; Foster-Brown, L.; Pognan, F.; Gadient, R.; Jacobs, R.T.; Zacco, A.; Greenberg, B.; Ciaccio, P.J. Modulation of notch processing by gamma-secretase inhibitors causes intestinal goblet cell metaplasia and induction of genes known to specify gut secretory lineage differentiation. Toxicol. Sci. 2004, 82, 341–358. [Google Scholar] [CrossRef]
- Wong, G.T.; Manfra, D.; Poulet, F.M.; Zhang, Q.; Josien, H.; Bara, T.; Engstrom, L.; Pinzon-Ortiz, M.; Fine, J.S.; Lee, H.J.; et al. Chronic treatment with the gamma-secretase inhibitor LY-411,575 inhibits beta-amyloid peptide production and alters lymphopoiesis and intestinal cell differentiation. J. Biol. Chem. 2004, 279, 12876–12882. [Google Scholar] [CrossRef] [Green Version]
- Jensen, J.; Pedersen, E.E.; Galante, P.; Hald, J.; Heller, R.S.; Ishibashi, M.; Kageyama, R.; Guillemot, F.; Serup, P.; Madsen, O.D. Control of endodermal endocrine development by Hes-1. Nat. Genet. 2000, 24, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Schoenborn, A.A.; von Furstenberg, R.J.; Valsaraj, S.; Hussain, F.S.; Stein, M.; Shanahan, M.T.; Henning, S.J.; Gulati, A.S. The enteric microbiota regulates jejunal Paneth cell number and function without impacting intestinal stem cells. Gut Microbes 2019, 10, 45–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lueschow, S.R.; McElroy, S.J. The Paneth cell: The curator and defender of the immature small intestine. Front. Immunol. 2020, 11, 587. [Google Scholar] [CrossRef] [PubMed]
- Pellegrinet, L.; Rodilla, V.; Liu, Z.; Chen, S.; Koch, U.; Espinosa, L.; Kaestner, K.H.; Kopan, R.; Lewis, J.; Radtke, F. Dll1- and dll4-mediated notch signaling are required for homeostasis of intestinal stem cells. Gastroenterology 2011, 140, 1230–1240.e7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shroyer, N.F.; Wong, M.H. BMP signaling in the intestine: Cross-talk is key. Gastroenterology 2007, 133, 1035–1038. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Wu, C. The relationship between intestinal goblet cells and the immune response. Biosci. Rep. 2020, 40, BSR20201471. [Google Scholar] [CrossRef] [PubMed]
- Miron, N.; Cristea, V. Enterocytes: Active cells in tolerance to food and microbial antigens in the gut. Clin. Exp. Immunol. 2012, 167, 405–412. [Google Scholar] [CrossRef]
- Yu, Y.; Yang, W.; Li, Y.; Cong, Y. Enteroendocrine cells: Sensing gut microbiota and regulating inflammatory bowel diseases. Inflamm. Bowel. Dis. 2020, 26, 11–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gerbe, F.; Jay, P. Intestinal tuft cells: Epithelial sentinels linking luminal cues to the immune system. Mucosal Immunol. 2016, 9, 1353–1359. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.; McKinley, E.T.; von Moltke, J.; Coffey, R.J.; Lau, K.S. Interpreting heterogeneity in intestinal tuft cell structure and function. J. Clin. Investig. 2018, 128, 1711–1719. [Google Scholar] [CrossRef] [PubMed]
- Dillon, A.; Lo, D.D. M Cells: Intelligent engineering of mucosal immune surveillance. Front. Immunol. 2019, 10, 1499. [Google Scholar] [CrossRef] [PubMed]
- Karam, S.M. Lineage commitment and maturation of epithelial cells in the gut. Front. Biosci. 1999, 4, D286–D298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schonhoff, S.E.; Giel-Moloney, M.; Leiter, A.B. Minireview: Development and differentiation of gut endocrine cells. Endocrinology 2004, 145, 2639–2644. [Google Scholar] [CrossRef] [Green Version]
- McGuckin, M.A.; Eri, R.; Simms, L.A.; Florin, T.H.; Radford-Smith, G. Intestinal barrier dysfunction in inflammatory bowel diseases. Inflamm. Bowel Dis. 2009, 15, 100–113. [Google Scholar] [CrossRef] [Green Version]
- Johansson, M.E.; Sjövall, H.; Hansson, G.C. The gastrointestinal mucus system in health and disease. Nat. Rev. Gastroenterol. Hepatol. 2013, 10, 352–361. [Google Scholar] [CrossRef] [Green Version]
- Celli, J.P.; Turner, B.S.; Afdhal, N.H.; Keates, S.; Ghiran, I.; Kelly, C.P.; Ewoldt, R.H.; McKinley, G.H.; So, P.; Erramilli, S.; et al. Helicobacter pylori moves through mucus by reducing mucin viscoelasticity. Proc. Natl. Acad. Sci. USA 2009, 106, 14321–14326. [Google Scholar] [CrossRef] [Green Version]
- Reunanen, J.; Kainulainen, V.; Huuskonen, L.; Ottman, N.; Belzer, C.; Huhtinen, H.; de Vos, W.M.; Satokari, R. Akkermansia muciniphila adheres to enterocytes and strengthens the integrity of the epithelial cell layer. Appl. Environ. Microbiol. 2015, 81, 3655–3662. [Google Scholar] [CrossRef] [Green Version]
- Schnupf, P.; Gaboriau-Routhiau, V.; Cerf-Bensussan, N. Host interactions with Segmented Filamentous Bacteria: An unusual trade-off that drives the post-natal maturation of the gut immune system. Semin. Immunol. 2013, 25, 342–351. [Google Scholar] [CrossRef]
- Swidsinski, A.; Sydora, B.C.; Doerffel, Y.; Loening-Baucke, V.; Vaneechoutte, M.; Lupicki, M.; Scholze, J.; Lochs, H.; Dieleman, L.A. Viscosity gradient within the mucus layer determines the mucosal barrier function and the spatial organization of the intestinal microbiota. Inflamm. Bowel Dis. 2007, 13, 963–970. [Google Scholar] [CrossRef]
- Garrett, W.S.; Gordon, J.I.; Glimcher, L.H. Homeostasis and inflammation in the intestine. Cell 2010, 140, 859–870. [Google Scholar] [CrossRef] [Green Version]
- Dethlefsen, L.; McFall-Ngai, M.; Relman, D.A. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature 2007, 449, 811–818. [Google Scholar] [CrossRef] [PubMed]
- Van der Sluis, M.; De Koning, B.A.; De Bruijn, A.C.; Velcich, A.; Meijerink, J.P.; Van Goudoever, J.B.; Büller, H.A.; Dekker, J.; Van Seuningen, I.; Renes, I.B.; et al. Muc2-deficient mice spontaneously develop colitis, indicating that MUC2 is critical for colonic protection. Gastroenterology 2006, 131, 117–129. [Google Scholar] [CrossRef] [PubMed]
- Newmark, H.L.; Lipkin, M.; Maheshwari, N. Colonic hyperplasia and hyperproliferation induced by a nutritional stress diet with four components of Western-style diet. J. Natl. Cancer Inst. 1990, 82, 491–496. [Google Scholar] [CrossRef] [PubMed]
- Newmark, H.L.; Lipkin, M.; Maheshwari, N. Colonic hyperproliferation induced in rats and mice by nutritional-stress diets containing four components of a human Western-style diet (series 2). Am. J. Clin. Nutr. 1991, 54 (Suppl. S1), 209S–214S. [Google Scholar] [CrossRef]
- Li, W.; Zimmerman, S.E.; Peregrina, K.; Houston, M.; Mayoral, J.; Zhang, J.; Maqbool, S.; Zhang, Z.; Cai, Y.; Ye, K.; et al. The nutritional environment determines which and how intestinal stem cells contribute to homeostasis and tumorigenesis. Carcinogenesis 2019, 40, 937–946. [Google Scholar] [CrossRef]
- Yang, K.; Kurihara, N.; Fan, K.; Newmark, H.; Rigas, B.; Bancroft, L.; Corner, G.; Livote, E.; Lesser, M.; Edelmann, W.; et al. Dietary induction of colonic tumors in a mouse model of sporadic colon cancer. Cancer Res. 2008, 68, 7803–7810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, D.; Peregrina, K.; Dhima, E.; Lin, E.Y.; Mariadason, J.M.; Augenlicht, L.H. Paneth cell marker expression in intestinal villi and colon crypts characterizes dietary induced risk for mouse sporadic intestinal cancer. Proc. Natl. Acad. Sci. USA 2011, 108, 10272–10277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peregrina, K.; Houston, M.; Daroqui, C.; Dhima, E.; Sellers, R.S.; Augenlicht, L.H. Vitamin D is a determinant of mouse intestinal Lgr5 stem cell functions. Carcinogenesis 2015, 36, 25–31. [Google Scholar] [CrossRef] [Green Version]
- Cheng, C.W.; Biton, M.; Haber, A.L.; Gunduz, N.; Eng, G.; Gaynor, L.T.; Tripathi, S.; Calibasi-Kocal, G.; Rickelt, S.; Butty, V.L.; et al. Ketone body signaling mediates intestinal stem cell homeostasis and adaptation to diet. Cell 2019, 178, 1115–1131.e15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alsharairi, N.A. the role of short-chain fatty acids in the interplay between a very low-calorie ketogenic diet and the infant gut microbiota and its therapeutic implications for reduced asthma. Int. J. Mol. Sci. 2020, 21, 9580. [Google Scholar] [CrossRef] [PubMed]
- Paoli, A.; Mancin, L.; Bianco, A.; Thomas, E.; Mota, J.F.; Piccini, F. Ketogenic diet and microbiota: Friends or enemies? Genes 2019, 10, 534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hennessy, A.A.; Ross, R.P.; Devery, R.; Stanton, C. The health promoting properties of the conjugated isomers of α-linolenic acid. Lipids 2011, 46, 105–119. [Google Scholar] [CrossRef]
- Yuan, G.F.; Chen, X.E.; Li, D. Conjugated linolenic acids and their bioactivities: A review. Food Funct. 2014, 5, 1360–1368. [Google Scholar] [CrossRef]
- Fontes, A.L.; Pimentel, L.L.; Simões, C.D.; Gomes, A.M.P.; Rodríguez-Alcalá, L.M. Evidences and perspectives in the utilization of CLNA isomers as bioactive compounds in foods. Crit. Rev. Food Sci. Nutr. 2017, 57, 2611–2622. [Google Scholar] [CrossRef]
- Benoit, B.; Bruno, J.; Kayal, F.; Estienne, M.; Debard, C.; Ducroc, R.; Plaisancié, P. Saturated and unsaturated fatty acids differently modulate colonic goblet cells in vitro and in rat pups. J. Nutr. 2015, 145, 1754–1762. [Google Scholar] [CrossRef] [Green Version]
- Todorov, H.; Kollar, B.; Bayer, F.; Brandão, I.; Mann, A.; Mohr, J.; Pontarollo, G.; Formes, H.; Stauber, R.; Kittner, J.M.; et al. α-Linolenic acid-rich diet influences microbiota composition and villus morphology of the mouse small intestine. Nutrients 2020, 12, 732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, S.; Xia, Y.; Zhu, G.; Yan, J.; Tan, C.; Deng, B.; Deng, J.; Yin, Y.; Ren, W. Glutamine supplementation improves intestinal cell proliferation and stem cell differentiation in weanling mice. Food Nutr. Res. 2018, 62. [Google Scholar] [CrossRef]
- Moore, S.R.; Guedes, M.M.; Costa, T.B.; Vallance., J.; Maier, E.A.; Betz, K.J.; Aihara, E.; Mahe, M.M.; Lima, A.A.; Oriá, R.B.; et al. Glutamine and alanyl-glutamine promote crypt expansion and mTOR signaling in murine enteroids. Am. J. Physiol. Gastrointest. Liver Physiol. 2015, 308, G831–G839. [Google Scholar] [CrossRef] [Green Version]
- Scheppach, W.; Loges, C.; Bartram, P.; Christl, S.U.; Richter, F.; Dusel, G.; Stehle, P.; Fuerst, P.; Kasper, H. Effect of free glutamine and alanyl-glutamine dipeptide on mucosal proliferation of the human ileum and colon. Gastroenterology 1994, 107, 429–434. [Google Scholar] [CrossRef]
- Beaufrère, A.M.; Neveux, N.; Patureau Mirand, P.; Buffière, C.; Marceau, G.; Sapin, V.; Cynober, L.; Meydinal-Denis, D. Long-term intermittent glutamine supplementation repairs intestinal damage (structure and functional mass) with advanced age: Assessment with plasma citrulline in a rodent model. J. Nutr. Health Aging. 2014, 18, 814–819. [Google Scholar] [CrossRef] [PubMed]
- Ren, W.; Duan, J.; Yin, J.; Liu, G.; Cao, Z.; Xiong, X.; Chen, S.; Li, T.; Yin, Y.; Hou, Y.; et al. Dietary L-glutamine supplementation modulates microbial community and activates innate immunity in the mouse intestine. Amino Acids. 2014, 46, 2403–2413. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Tseng, S.H.; Yao, C.L.; Li, C.; Tsai, Y.H. Distinct effects of growth hormone and glutamine on activation of intestinal stem cells. JPEN J. Parenter. Enter. Nutr. 2018, 42, 642–651. [Google Scholar] [CrossRef]
- Liu, G.; Ren, W.; Fang, J.; Hu, C.A.; Guan, G.; Al-Dhabi, N.A.; Yin, J.; Duraipandiyan, V.; Chen, S.; Peng, Y.; et al. L-Glutamine and L-arginine protect against enterotoxigenic Escherichia coli infection via intestinal innate immunity in mice. Amino Acids. 2017, 49, 1945–1954. [Google Scholar] [CrossRef] [PubMed]
- McKee, C.M.; Penno, M.B.; Cowman, M.; Burdick, M.D.; Strieter, R.M.; Bao, C.; Noble, P.W. Hyaluronan (HA) fragments induce chemokine gene expression in alveolar macrophages. The role of HA size and CD44. J. Clin. Investig. 1996, 98, 2403–2413. [Google Scholar] [CrossRef] [Green Version]
- McKee, C.M.; Lowenstein, C.J.; Horton, M.R.; Wu, J.; Bao, C.; Chin, B.Y.; Choi, A.M.; Noble, P.W. Hyaluronan fragments induce nitric-oxide synthase in murine macrophages through a nuclear factor kappaB-dependent mechanism. J. Biol. Chem. 1997, 272, 8013–8018. [Google Scholar] [CrossRef] [Green Version]
- Majors, A.K.; Austin, R.C.; de la Motte, C.A.; Pyeritz, R.E.; Hascall, V.C.; Kessler, S.P.; Sen, G.; Strong, S.A. Endoplasmic reticulum stress induces hyaluronan deposition and leukocyte adhesion. J. Biol. Chem. 2003, 278, 47223–47231. [Google Scholar] [CrossRef] [Green Version]
- Zheng, L.; Riehl, T.E.; Stenson, W.F. Regulation of colonic epithelial repair in mice by Toll-like receptors and hyaluronic acid. Gastroenterology 2009, 137, 2041–2051. [Google Scholar] [CrossRef] [Green Version]
- Riehl, T.E.; Ee, X.; Stenson, W.F. Hyaluronic acid regulates normal intestinal and colonic growth in mice. Am. J. Physiol. Gastrointest. Liver Physiol. 2012, 303, G377–G388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yilmaz, Ö.H.; Katajisto, P.; Lamming, D.W.; Gültekin, Y.; Bauer-Rowe, K.E.; Sengupta, S.; Birsoy, K.; Dursun, A.; Yilmaz, V.O.; Selig, M.; et al. mTORC1 in the Paneth cell niche couples intestinal stem-cell function to calorie intake. Nature 2012, 486, 490–495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richmond, C.A.; Shah, M.S.; Deary, L.T.; Trotier, D.C.; Thomas, H.; Ambruzs, D.M.; Jiang, L.; Whiles, B.B.; Rickner, H.D.; Montgomery, R.K.; et al. Dormant intestinal stem cells are regulated by PTEN and nutritional status. Cell Rep. 2015, 13, 2403–2411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burgueño, J.F.; Abreu, M.T. Epithelial Toll-like receptors and their role in gut homeostasis and disease. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 263–278. [Google Scholar] [CrossRef] [PubMed]
- Fukata, M.; Vamadevan, A.S.; Abreu, M.T. Toll-like receptors (TLRs) and Nod-like receptors (NLRs) in inflammatory disorders. Semin. Immunol. 2009, 21, 242–253. [Google Scholar] [CrossRef] [PubMed]
- Abreu, M.T. Toll-like receptor signalling in the intestinal epithelium: How bacterial recognition shapes intestinal function. Nat. Rev. Immunol. 2010, 10, 131–144. [Google Scholar] [CrossRef] [PubMed]
- Bogunovic, M.; Davé, S.H.; Tilstra, J.S.; Chang, D.T.W.; Harpaz, N.; Xiong, H.; Mayer, L.F.; Plevy, S.E. Enteroendocrine cells express funcitonal Toll-like receptors. Am. J. Physiol. Gastrointest. Liver Physiol. 2007, 292, G1770–G1783. [Google Scholar] [CrossRef] [PubMed]
- Cetin, S.; Ford, H.R.; Sysko, L.R.; Agarwal, C.; Wang, J.; Neal, M.D.; Baty, C.; Apodaca, G.; Hackam, D.J. Endotoxin inhibits intestinal epithelial restitution through activation of Rho-GTPase and increased focal adhesions. J. Biol. Chem. 2004, 279, 24592–24600. [Google Scholar] [CrossRef] [Green Version]
- Ikeda, H.; Sasaki, M.; Ishikawa, A.; Sato, Y.; Harada, K.; Zen, Y.; Kazumori, H.; Nakanuma, Y. Interaction of Toll-like receptors with bacterial components induces expression of CDX2 and MUC2 in rat biliary epithelium in vivo and in culture. Lab. Investig. 2007, 87, 559–571. [Google Scholar] [CrossRef] [PubMed]
- Yi, H.; Patel, A.K.; Sodhi, C.P.; Hackam, D.J.; Hackam, A.S. Novel role for the innate immune receptor Toll-like receptor 4 (TLR4) in the regulation of the Wnt signaling pathway and photoreceptor apoptosis. PLoS ONE 2012, 7, e36560. [Google Scholar] [CrossRef] [Green Version]
- Sodhi, C.P.; Shi, X.H.; Richardson, W.M.; Grant, Z.S.; Shapiro, R.A.; Prindle, T., Jr.; Branca, M.; Russo, A.; Gribar, S.C.; Ma, C.; et al. Toll-like receptor-4 inhibits enterocyte proliferation via impaired beta-catenin signaling in necrotizing enterocolitis. Gastroenterology 2010, 138, 185–196. [Google Scholar] [CrossRef] [Green Version]
- Sodhi, C.P.; Neal, M.D.; Siggers, R.; Sho, S.; Ma, C.; Branca, M.F.; Prindle, T., Jr.; Russo, A.M.; Afrazi, A.; Good, M.; et al. Intestinal epithelial Toll-like receptor 4 regulates goblet cell development and is required for necrotizing enterocolitis in mice. Gastroenterology 2012, 143, 708–718.e5. [Google Scholar] [CrossRef] [Green Version]
- Rakoff-Nahoum, S.; Paglino, J.; Eslami-Varzaneh, F.; Edberg, S.; Medzhitov, R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell 2004, 118, 229–241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cario, E.; Gerken, G.; Podolsky, D.K. Toll-like receptor 2 controls mucosal inflammation by regulating epithelial barrier function. Gastroenterology 2007, 132, 1359–1374. [Google Scholar] [CrossRef]
- Sailaja, B.S.; He, X.C.; Li, L. The regulatory niche of intestinal stem cells. J. Physiol. 2016, 594, 4827–4836. [Google Scholar] [CrossRef] [Green Version]
- Lindemans, C.A.; Calafiore, M.; Mertelsmann, A.M.; O’Connor, M.H.; Dudakov, J.A.; Jenq, R.R.; Velardi, E.; Young, L.F.; Smith, O.M.; Lawrence, G.; et al. Interleukin-22 promotes intestinal-stem-cell-mediated epithelial regeneration. Nature 2015, 528, 560–564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mizoguchi, A. Healing of intestinal inflammation by IL-22. Inflamm. Bowel Dis. 2012, 18, 1777–1784. [Google Scholar] [CrossRef] [PubMed]
- Lucey, D.R.; Clerici, M.; Shearer, G.M. Type 1 and type 2 cytokine dysregulation in human infectious, neoplastic, and inflammatory diseases. Clin. Microbiol. Rev. 1996, 9, 532–562. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.J.; Kim, D.; Pyo, K.H.; Kim, M.K.; Kim, H.J.; Chai, J.Y.; Shin, E.H. STAT6 expression and IL-13 production in association with goblet cell hyperplasia and worm expulsion of Gymnophalloides seoi from C57BL/6 mice. Korean J. Parasitol. 2013, 51, 589–594. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.M.; Patel, P.S.; Bao, K.; Danhorn, T.; O’Connor, B.P.; Reinhardt, R.L. BATF acts as an essential regulator of IL-25-responsive migratory ILC2 cell fate and function. Sci. Immunol. 2020, 5, eaay3994. [Google Scholar] [CrossRef] [PubMed]
- Marillier, R.G.; Michels, C.; Smith, E.M.; Fick, L.C.; Leeto, M.; Dewals, B.; Horsnell, W.G.; Brombacher, F. IL-4/IL-13 independent goblet cell hyperplasia in experimental helminth infections. BMC Immunol. 2008, 9, 11. [Google Scholar] [CrossRef] [Green Version]
- Kuhn, K.A.; Manieri, N.A.; Liu, T.C.; Stappenbeck, T.S. IL-6 stimulates intestinal epithelial proliferation and repair after injury. PLoS ONE 2014, 9, e114195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Capaldo, C.T.; Beeman, N.; Hilgarth, R.S.; Nava, P.; Louis, N.A.; Naschberger, E.; Stürzl, M.; Parkos, C.A.; Nusrat, A. IFN-γ and TNF-α-induced GBP-1 inhibits epithelial cell proliferation through suppression of β-catenin/TCF signaling. Mucosal Immunol. 2012, 5, 681–690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akcora, D.; Huynh, D.; Lightowler, S.; Germann, M.; Robine, S.; de May, J.R.; Pollard, J.W.; Stanley, E.R.; Malaterre, J.; Ramsay, R.G. The CSF-1 receptor fashions the intestinal stem cell niche. Stem Cell Res. 2013, 10, 203–212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saha, S.; Aranda, E.; Hayakawa, Y.; Bhanja, P.; Atay, S.; Brodin, N.P.; Li, J.; Asfaha, S.; Liu, L.; Tailor, Y.; et al. Macrophage-derived extracellular vesicle-packaged WNTs rescue intestinal stem cells and enhance survival after radiation injury. Nat. Commun. 2016, 7, 13096. [Google Scholar] [CrossRef]
- Nagashima, R.; Maeda, K.; Imai, Y.; Takahashi, T. Lamina propria macrophages in the human gastrointestinal mucosa: Their distribution, immunohistological phenotype, and function. J. Histochem. Cytochem. 1996, 44, 721–731. [Google Scholar] [CrossRef] [PubMed]
- Kang, B.; Alvarado, L.J.; Kim, T.; Lehmann, M.L.; Cho, H.; He, J.; Li, P.; Kim, B.H.; Larochelle, A.; Kelsall, B.L. Commensal microbiota drive the functional diversification of colon macrophages. Mucosal Immunol. 2020, 13, 216–229. [Google Scholar] [CrossRef] [Green Version]
- Chikina, A.S.; Nadalin, F.; Maurin, M.; San-Roman, M.; Thomas-Bonafos, T.; Li, X.V.; Lameiras, S.; Baulande, S.; Henri, S.; Malissen, B.; et al. Macrophages maintain epithelium integrity by limiting fungal product absorption. Cell 2020, 183, 411–428.e16. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Chou, K.; Fuchs, E.; Havran, W.L.; Boismenu, R. Protection of the intestinal mucosa by intraepithelial gamma delta T cells. Proc. Natl. Acad. Sci. USA 2002, 99, 14338–14343. [Google Scholar] [CrossRef] [Green Version]
- Hills, R.D., Jr.; Pontefract, B.A.; Mishcon, H.R.; Black, C.A.; Sutton, S.C.; Theberge, C.R. Gut microbiome: Profound implications for diet and disease. Nutrients 2019, 11, 1613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Francescangeli, F.; De Angelis, M.L.; Zeuner, A. Dietary factors in the control of gut homeostasis, intestinal stem cells, and colorectal cancer. Nutrients 2019, 11, 2936. [Google Scholar] [CrossRef] [Green Version]
- Leeming, E.R.; Johnson, A.J.; Spector, T.D.; Le Roy, C.I. Effect of diet on the gut microbiota: Rethinking intervention duration. Nutrients 2019, 11, 2862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Husted, A.S.; Trauelsen, M.; Rudenko, O.; Hjorth, S.A.; Schwartz, T.W. GPCR-mediated signaling of metabolites. Cell Metab. 2017, 25, 777–796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Topping, D.L.; Clifton, P.M. Short-chain fatty acids and human colonic function: Roles of resistant starch and nonstarch polysaccharides. Physiol. Rev. 2001, 81, 1031–1064. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.M.; de Souza, R.; Kendall, C.W.; Emam, A.; Jenkins, D.J. Colonic health: Fermentation and short chain fatty acids. J. Clin. Gastroenterol. 2006, 40, 235–243. [Google Scholar] [CrossRef] [PubMed]
- De Vadder, F.; Kovatcheva-Datchary, P.; Goncalves, D.; Vinera, J.; Zitoun, C.; Duchampt, A.; Bäckhed, F.; Mithieux, G. Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell 2014, 156, 84–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fung, K.Y.; Brierley, G.V.; Henderson, S.; Hoffmann, P.; McColl, S.R.; Lockett, T.; Head, R.; Cosgrove, L. Butyrate-induced apoptosis in HCT116 colorectal cancer cells includes induction of a cell stress response. J. Proteome Res. 2011, 10, 1860–1869. [Google Scholar] [CrossRef] [PubMed]
- Byndloss, M.X.; Olsan, E.E.; Rivera-Chávez, F.; Tiffany, C.R.; Cevallos, S.A.; Lokken, K.L.; Torres, T.P.; Byndloss, A.J.; Faber, F.; Gao, Y.; et al. Microbiota-activated PPAR-γ signaling inhibits dysbiotic Enterobacteriaceae expansion. Science 2017, 357, 570–575. [Google Scholar] [CrossRef]
- Koh, A.; De Vadder, F.; Kovatcheva-Datchary, P.; Bäckhed, F. From dietary fiber to host physiology: Short-chain fatty acids as key Bbcterial metabolites. Cell 2016, 165, 1332–1345. [Google Scholar] [CrossRef] [Green Version]
- Frost, G.; Sleeth, M.L.; Sahuri-Arisoylu, M.; Lizarbe, B.; Cerdan, S.; Brody, L.; Anastasovska, J.; Ghourab, S.; Hankir, M.; Zhang, S.; et al. The short-chain fatty acid acetate reduces appetite via a central homeostatic mechanism. Nat. Commun. 2014, 5, 3611. [Google Scholar] [CrossRef] [Green Version]
- O’Keefe, S.J. Nutrition and colonic health: The critical role of the microbiota. Curr. Opin. Gastroenterol. 2008, 24, 51–58. [Google Scholar] [CrossRef]
- Zarrinpar, A.; Chaix, A.; Xu, Z.Z.; Chang, M.W.; Marotz, C.A.; Saghatelian, A.; Knight, R.; Panda, S. Antibiotic-induced microbiome depletion alters metabolic homeostasis by affecting gut signaling and colonic metabolism. Nat. Commun. 2018, 9, 2872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, H.; Sun, Y.; Wu, Y.; Luan, B.; Wang, Y.; Qu, B.; Pei, G. Identification of beta-arrestin2 as a G protein-coupled receptor-stimulated regulator of NF-kappaB pathways. Mol. Cell. 2004, 14, 303–317. [Google Scholar] [CrossRef]
- Kim, M.H.; Kang, S.G.; Park, J.H.; Yanagisawa, M.; Kim, C.H. Short-chain fatty acids activate GPR41 and GPR43 on intestinal epithelial cells to promote inflammatory responses in mice. Gastroenterology 2013, 145, 396–406.10. [Google Scholar] [CrossRef]
- Macia, L.; Tan, J.; Vieira, A.T.; Leach, K.; Stanley, D.; Luong, S.; Maruya, M.; Ian McKenzie, C.; Hijikata, A.; Wong, C.; et al. Metabolite-sensing receptors GPR43 and GPR109A facilitate dietary fibre-induced gut homeostasis through regulation of the inflammasome. Nat. Commun. 2015, 6, 6734. [Google Scholar] [CrossRef] [Green Version]
- Gaudier, E.; Jarry, A.; Blottière, H.M.; de Coppet, P.; Buisine, M.P.; Aubert, J.P.; Laboisse, C.; Cherbut, C.; Hoebler, C. Butyrate specifically modulates MUC gene expression in intestinal epithelial goblet cells deprived of glucose. Am. J. Physiol. Gastrointest. Liver Physiol. 2004, 287, G1168–G1174. [Google Scholar] [CrossRef] [Green Version]
- Willemsen, L.E.; Koetsier, M.A.; van Deventer, S.J.; van Tol, E.A. Short chain fatty acids stimulate epithelial mucin 2 expression through differential effects on prostaglandin E(1) and E(2) production by intestinal myofibroblasts. Gut 2003, 52, 1442–1447. [Google Scholar] [CrossRef] [Green Version]
- Alex, S.; Lange, K.; Amolo, T.; Grinstead, J.S.; Haakonsson, A.K.; Szalowska, E.; Koppen, A.; Mudde, K.; Haenen, D.; Al-Lahham, S.; et al. Short-chain fatty acids stimulate angiopoietin-like 4 synthesis in human colon adenocarcinoma cells by activating peroxisome proliferator-activated receptor γ. Mol. Cell. Biol. 2013, 33, 1303–1316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Varga, T.; Czimmerer, Z.; Nagy, L. PPARs are a unique set of fatty acid regulated transcription factors controlling both lipid metabolism and inflammation. Biochim. Biophys. Acta 2011, 1812, 1007–1022. [Google Scholar] [CrossRef]
- Litvak, Y.; Byndloss, M.X.; Bäumler, A.J. Colonocyte metabolism shapes the gut microbiota. Science 2018, 362, eaat9076. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.S.; Kim, T.Y.; Kim, Y.; Lee, S.H.; Kim, S.; Kang, S.W.; Yang, J.Y.; Baek, I.J.; Sung, Y.H.; Park, Y.Y.; et al. Microbiota-Derived Lactate Accelerates Intestinal Stem-Cell-Mediated Epithelial Development. Cell Host Microbe 2018, 24, 833–846.e6. [Google Scholar] [CrossRef] [Green Version]
- Swanson, P.A., 2nd; Kumar, A.; Samarin, S.; Vijay-Kumar, M.; Kundu, K.; Murthy, N.; Hansen, J.; Nusrat, A.; Neish, A.S. Enteric commensal bacteria potentiate epithelial restitution via reactive oxygen species-mediated inactivation of focal adhesion kinase phosphatases. Proc. Natl. Acad. Sci. USA 2011, 108, 8803–8808. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levenson, S.M.; Tennant, B. Some metabolic and nutritional studies with germfree animals. Fed Proc. 1963, 22, 109–119. [Google Scholar] [PubMed]
- Gordon, H.A. Morphological and physiological characterization of germfree life. Ann. N. Y. Acad. Sci. 1959, 78, 208–220. [Google Scholar] [CrossRef] [PubMed]
- Meslin, J.C.; Sacquet, E.; Guenet, J.L. Action de la flore bactérienne sur la morphologie et la surface de la muqueuse de l’intestin grêle du rat [Action of bacterial flora on the morphology and surface mucus of the small intestine of the rat]. Ann. Biol. Anim. Biochim. Biophys. 1973, 13, 203–214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lesher, S.; Walburg, H.E., Jr.; Sacher, G.A., Jr. Generation cycle in the duodenal crypt cells of germ-free and conventional mice. Nature 1964, 202, 884–886. [Google Scholar] [CrossRef]
- Abrams, G.D.; Bauer, H.; Sprinz, H. Influence of the normal flora on mucosal morphology and cellular renewal in the ileum. A comparison of germ-free and conventional mice. Lab. Investig. 1963, 12, 355–364. [Google Scholar]
- Glaister, J.R. Light, fluorescence and electron microscopic studies of lymphoid cells in the small intestinal epithelium of mice. Int. Arch. Allergy Appl. Immunol. 1973, 45, 854–867. [Google Scholar] [CrossRef]
- Wostmann, B.; Bruckner-Kardoss, E. Development of cecal distention in germ-free baby rats. Am. J. Physiol. 1959, 197, 1345–1346. [Google Scholar] [CrossRef] [Green Version]
- Hudson, J.A.; Luckey, T.D. Bacteria induced morphologic changes. Proc. Soc. Exp. Biol. Med. 1964, 116, 628–631. [Google Scholar] [CrossRef]
- Coates, M.E.; Gordon, H.A.; Wostmann, B.S. The Germ-Free Animal in Research; Coates, M.E., Ed.; Academic Press: London, UK; New York, NY, USA, 1968. [Google Scholar]
- Jervis, H.R.; Biggers, D.C. Mucosal enzymes of the cecum of conventional and germfree mice. Anat. Rec. 1964, 148, 591–597. [Google Scholar] [CrossRef]
- Meslin, J.C.; Fontaine, N.; Andrieux, C. Variation of mucin distribution in the rat intestine, caecum and colon: Effect of the bacterial flora. Comp. Biochem. Physiol. A. Mol. Integr. Physiol. 1999, 123, 235–239. [Google Scholar] [CrossRef]
- Szentkuti, L.; Riedesel, H.; Enss, M.L.; Gaertner, K.; von Engelhardt, W. Pre-epithelial mucus layer in the colon of conventional and germ-free rats. Histochem. J. 1990, 22, 491–497. [Google Scholar] [CrossRef] [PubMed]
- Enss, M.L.; Grosse-Siestrup, H.; Schmidt-Wittig, U.; Gärtner, K. Changes in colonic mucins of germfree rats in response to the introduction of a “normal” rat microbial flora. Rat colonic mucin. J. Exp. Anim. Sci. 1992, 35, 110–119. [Google Scholar] [PubMed]
- Kandori, H.; Hirayama, K.; Takeda, M.; Doi, K. Histochemical, lectin-histochemical and morphometrical characteristics of intestinal goblet cells of germfree and conventional mice. Exp. Anim. 1996, 45, 155–160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petersson, J.; Schreiber, O.; Hansson, G.C.; Gendler, S.J.; Velcich, A.; Lundberg, J.O.; Roos, S.; Holm, L.; Phillipson, M. Importance and regulation of the colonic mucus barrier in a mouse model of colitis. Am. J. Physiol. Gastrointest. Liver Physiol. 2011, 300, G327–G333. [Google Scholar] [CrossRef] [Green Version]
- Preidis, G.A.; Saulnier, D.M.; Blutt, S.E.; Mistretta, T.A.; Riehle, K.P.; Major, A.M.; Venable, S.F.; Finegold, M.J.; Petrosino, J.F.; Conner, M.E.; et al. Probiotics stimulate enterocyte migration and microbial diversity in the neonatal mouse intestine. FASEB J. 2012, 26, 1960–1969. [Google Scholar] [CrossRef] [Green Version]
- Cifarelli, V.; Eichmann, A. The intestinal lymphatic system: Functions and metabolic implications. Cell. Mol. Gastroenterol. Hepatol. 2019, 7, 503–513. [Google Scholar] [CrossRef] [Green Version]
- Nurmi, H.; Saharinen, P.; Zarkada, G.; Zheng, W.; Robciuc, M.R.; Alitalo, K. VEGF-C is required for intestinal lymphatic vessel maintenance and lipid absorption. EMBO Mol. Med. 2015, 7, 1418–1425. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Zarkada, G.; Han, J.; Li, J.; Dubrac, A.; Ola, R.; Genet, G.; Boyé, K.; Michon, P.; Künzel, S.E.; et al. Lacteal junction zippering protects against diet-induced obesity. Science 2018, 361, 599–603. [Google Scholar] [CrossRef] [Green Version]
- Danese, S.; Scaldaferri, F.; Vetrano, S.; Stefanelli, T.; Graziani, C.; Repici, A.; Ricci, R.; Straface, G.; Sgambato, A.; Malesci, A.; et al. Critical role of the CD40 CD40-ligand pathway in regulating mucosal inflammation-driven angiogenesis in inflammatory bowel disease. Gut 2007, 56, 1248–1256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alkim, C.; Alkim, H.; Koksal, A.R.; Boga, S.; Sen, I. Angiogenesis in inflammatory bowel disease. Int. J. Inflam. 2015, 2015, 970890. [Google Scholar] [CrossRef] [Green Version]
- Myrsky, E.; Syrjänen, M.; Korponay-Szabo, I.R.; Mäki, M.; Kaukinen, K.; Lindfors, K. Altered small-bowel mucosal vascular network in untreated coeliac disease. Scand. J. Gastroenterol. 2009, 44, 162–167. [Google Scholar] [CrossRef]
- Park, H.; Kim, M.; Kwon, T.; Lim, D.Y.; Yu, R.; Sung, M.-K.; Lee, K.W.; Daily, J.W., 3rd; Park, J.H.Y. A gigh-fat diet increases angiogenesis, solid tumor growth, and lung metastasis of CT26 colon cancer cells in obesity-resistant BALB/c mice. Mol. Carcionog. 2012, 51, 869–880. [Google Scholar] [CrossRef] [PubMed]
- Khandagale, A.; Reinhardt, C. Gut microbiota—Architects of small intestinal capillaries. Front. Biosci. 2018, 23, 752–766. [Google Scholar] [CrossRef]
- Diaz-Miron, J.; Sun, R.; Choi, P.; Sommovilla, J.; Guo, J.; Erwin, C.R.; Mei, J.; Scott Worthen, G.; Warner, B.W. The effect of impaired angiogenesis on intestinal function following massive small bowel resection. J. Pediatr. Surg. 2015, 50, 948–953. [Google Scholar] [CrossRef] [Green Version]
- Monk, J.M.; Hou, T.Y.; Turk, H.F.; Weeks, B.; Wu, C.; McMurray, D.N.; Chapkin, R.S. Dietary n-3 polyunsaturated fatty acids (PUFA) decrease obesity-associated Th17 cell-mediated inflammation during colitis. PLoS ONE 2012, 7, e49739. [Google Scholar] [CrossRef] [Green Version]
- Johnson, A.M.; Costanzo, A.; Gareau, M.G.; Armando, A.M.; Quehenberger, O.; Jameson, J.M.; Olefsky, J.M. High fat diet causes depletion of intestinal eosinophils associated with intestinal permeability. PLoS ONE 2015, 10, e0122195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujiyama, Y.; Hokari, R.; Miura, S.; Watanabe, C.; Komoto, S.; Oyama, T.; Kurihara, C.; Nagata, H.; Hibi, T. Butter feeding enhances TNF-alpha production from macrophages and lymphocyte adherence in murine small intestinal microvessels. J. Gastroenterol. Hepatol. 2007, 22, 1838–1845. [Google Scholar] [CrossRef]
- Mouries, J.; Brescia, P.; Silvestri, A.; Spadoni, I.; Sorribas, M.; Wiest, R.; Mileti, E.; Galbiati, M.; Invernizzi, P.; Adorini, L.; et al. Microbiota-driven gut vascular barrier disruption is a prerequisite for non-alcoholic steatohepatitis development. J. Hepatol. 2019, 71, 1216–1228. [Google Scholar] [CrossRef] [Green Version]
- Battson, M.L.; Lee, D.M.; Jarrell, D.K.; Hou, S.; Ecton, K.E.; Weir, T.L.; Gentile, C.L. Suppression of gut dysbiosis reverses Western diet-induced vascular dysfunction. Am. J. Physiol. Endocrinol. Metab. 2018, 314, E468–E477. [Google Scholar] [CrossRef] [Green Version]
- Ibrahim, A.; Mbodji, K.; Hassan, A.; Aziz, M.; Boukhettala, N.; Coëffier, M.; Savoye, G.; Déchelotte, P.; Marion-Letellier, R. Anti-inflammatory and anti-angiogenic effect of long chain n-3 polyunsaturated fatty acids in intestinal microvascular endothelium. Clin. Nutr. 2011, 30, 678–687. [Google Scholar] [CrossRef]
- Ibrahim, A.; Aziz, M.; Hassan, A.; Mbodji, K.; Collasse, E.; Coëffier, M.; Bounoure, F.; Savoye, G.; Déchelotte, P.; Marion-Letellier, R. Dietary α-linolenic acid-rich formula reduces adhesion molecules in rats with experimental colitis. Nutrition 2012, 28, 799–802. [Google Scholar] [CrossRef]
- Martin, C.A.; Perrone, E.E.; Longshore, S.W.; Toste, P.; Bitter, K.; Nair, R.; Guo, J.; Erwin, C.R.; Warner, B.W. Intestinal resection induces angiogenesis within adapting intestinal villi. J. Pediatr. Surg. 2009, 44, 1077–1083, discussion 1083. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Onufer. E.J.; Czepielewski, R.; Seiler, K.M.; Erlich, E.; Courtney, C.M.; Bustos, A.; Randolph, G.J.; Warner, B.W. Lymphatic network remodeling after small bowel resection. J. Pediatr. Surg. 2019, 54, 1239–1244. [Google Scholar] [CrossRef]
- Kuroshima, S.; Sawa, Y.; Kawamoto, T.; Yamaoka, Y.; Notani, K.; Yoshida, S.; Inoue, N. Expression of Toll-like receptors 2 and 4 on human intestinal lymphatic vessels. Microvasc. Res. 2004, 67, 90–95. [Google Scholar] [CrossRef] [PubMed]
- Maaser, C.; Heidemann, J.; von Eiff, C.; Lugering, A.; Spahn, T.W.; Binion, D.G.; Domschke, W.; Lugering, N.; Kucharzik, T. Human intestinal microvascular endothelial cells express Toll-like receptor 5: A binding partner for bacterial flagellin. J. Immunol. 2004, 172, 5056–5062. [Google Scholar] [CrossRef] [Green Version]
- Schirbel, A.; Kessler, S.; Rieder, F.; West, G.; Rebert, N.; Asosingh, K.; McDonald, C.; Fiocchi, C. Pro-angiogenic activity of TLRs and NLRs: A novel link between gut microbiota and intestinal angiogenesis. Gastroenterology 2013, 144, 613–623.e9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ogawa, H.; Rafiee, P.; Heidemann, J.; Fisher, P.J.; Johnson, N.A.; Otterson, M.F.; Kalyanaraman, B.; Pritchard, K.A., Jr.; Binion, D.G. Mechanisms of endotoxin tolerance in human intestinal microvascular endothelial cells. J. Immunol. 2003, 170, 5956–5964. [Google Scholar] [CrossRef]
- Wagner, N.M.; Bierhansl, L.; Nöldge-Schomburg, G.; Vollmar, B.; Roesner, J.P. Toll-like receptor 2-blocking antibodies promote angiogenesis and induce ERK1/2 and AKT signaling via CXCR4 in endothelial cells. Arterioscler. Thromb. Vasc. Biol. 2013, 33, 1943–1951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rakoff-Nahoum, S.; Kong, Y.; Kleinstein, S.H.; Subramanian, S.; Ahern, P.P.; Gordon, J.I.; Medzhitov, R. Analysis of gene-environment interactions in postnatal development of the mammalian intestine. Proc. Natl. Acad. Sci. USA 2015, 112, 1929–1936. [Google Scholar] [CrossRef] [Green Version]
- Oviedo-Boyso, J.; Bravo-Patiño, A.; Baizabal-Aguirre, V.M. Collaborative action of Toll-like and NOD-like receptors as modulators of the inflammatory response to pathogenic bacteria. Mediators Inflamm. 2014, 2014, 432785. [Google Scholar] [CrossRef] [PubMed]
- Heidemann, J.; Ogawa, H.; Rafiee, P.; Lügering, N.; Maaser, C.; Domschke, W.; Binion, D.G.; Dwinell, M.B. Mucosal angiogenesis regulation by CXCR4 and its ligand CXCL12 expressed by human intestinal microvascular endothelial cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2004, 286, G1059–G1068. [Google Scholar] [CrossRef] [PubMed]
- De Schepper, S.; Verheijden, S.; Aguilera-Lizarraga, J.; Viola, M.F.; Boesmans, W.; Stakenborg, N.; Voytyuk, I.; Schmidt, I.; Boeckx, B.; Dierckx de Casterlé, I.; et al. Self-maintaining gut macrophages are essential for intestinal homeostasis. Cell 2018, 175, 400–415.e13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eder, P.; Lykowska-Szuber, L.; Iwanik, K.; Krela-Kazmierczak, I.; Stawczyk-Eder, K.; Majewski, P.; Linke, K.; Kay, E.W.; Wozniak, A. The influence of anti-TNF therapy on CD31 and VEGF expression in colonic mucosa of Crohn’s disease patients in relation to mucosal healing. Folia Histochem. Cytobiol. 2016, 54, 75–78. [Google Scholar] [CrossRef] [Green Version]
- Koon, H.W.; Shih, D.Q.; Hing, T.C.; Chen, J.; Ho, S.; Zhao, D.; Targan, S.R.; Pothoulakis, C. Substance P induces CCN1 expression via histone deacetylase activity in human colonic epithelial cells. Am. J. Pathol. 2011, 179, 2315–2326. [Google Scholar] [CrossRef]
- Zhu, G.; Huang, Q.; Huang, Y.; Zheng, W.; Hua, J.; Yang, S.; Zhuang, J.; Wang, J.; Ye, J. Lipopolysaccharide increases the release of VEGF-C that enhances cell motility and promotes lymphangiogenesis and lymphatic metastasis through the TLR4-NF-κB/JNK pathways in colorectal cancer. Oncotarget 2016, 7, 73711–73724. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ogawa, H.; Binion, D.G. Effect of enteric flora on inflammatory and angiogenic mechanisms in human intestinal microvascular endothelial cells. Front. Biosci. 2005, 10, 94–98. [Google Scholar] [CrossRef] [Green Version]
- Schnupf, P.; Gaboriau-Routhiau, V.; Sansonetti, P.J.; Cerf-Bensussan, N. Segmented filamentous bacteria, Th17 inducers and helpers in a hostile world. Curr. Opin. Microbiol. 2017, 35, 100–109. [Google Scholar] [CrossRef]
- Seiler, K.M.; Bajinting, A.; Alvarado, D.M.; Traore, M.A.; Binkley, M.M.; Goo, W.H.; Lanik, W.E.; Ou, J.; Ismail, U.; Iticovici, M.; et al. Patient-derived small intestinal myofibroblasts direct perfused, physiologically responsive capillary development in a microfluidic gut-on-a-chip model. Sci. Rep. 2020, 10, 3842. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schulte-Merker, S.; Sabine, A.; Petrova, T.V. Lymphatic vascular morphogenesis in development, physiology, and disease. J. Cell Biol. 2011, 193, 607–618. [Google Scholar] [CrossRef] [Green Version]
- Clavel, T.; Haller, D. Bacteria- and host-derived mechanisms to control intestinal epithelial cell homeostasis: Implications for chronic inflammation. Inflamm. Bowel Dis. 2007, 13, 1153–1164. [Google Scholar] [CrossRef]
- Faure, E.; Thomas, L.; Xu, H.; Medvedev, A.; Equils, O.; Arditi, M. Bacterial lipopolysaccharide and IFN-gamma induce Toll-like receptor 2 and Toll-like receptor 4 expression in human endothelial cells: Role of NF-kappa B activation. J. Immunol. 2001, 166, 2018–2024. [Google Scholar] [CrossRef] [PubMed]
- Davey, M.P.; Martin, T.M.; Planck, S.R.; Lee, J.; Zamora, D.; Rosenbaum, J.T. Human endothelial cells express NOD2/CARD15 and increase IL-6 secretion in response to muramyl dipeptide. Microvasc. Res. 2006, 71, 103–107. [Google Scholar] [CrossRef] [PubMed]
- Jeong, J.H.; Kim, K.; Lim, D.; Kim, K.H.; Kim, H.S.; Lee, S.; Song, J.H.; Moon, B.G.; Choy, H.E.; Park, S.C. Microvasculature remodeling in the mouse lower gut during inflammaging. Sci. Rep. 2017, 7, 39848. [Google Scholar] [CrossRef] [PubMed]
- Augustin, H.G.; Koh, G.Y.; Thurston, G.; Alitalo, K. Control of vascular morphogenesis and homeostasis through the angiopoietin-Tie system. Nat. Rev. Mol. Cell Biol. 2009, 10, 165–177. [Google Scholar] [CrossRef]
- Scaldaferri, F.; Vetrano, S.; Sans, M.; Arena, V.; Straface, G.; Stigliano, E.; Repici, A.; Sturm, A.; Malesci, A.; Panes, J.; et al. VEGF-A links angiogenesis and inflammation in inflammatory bowel disease pathogenesis. Gastroenterology 2009, 136, 585–595.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kayisoglu, O.; Weiss, F.; Niklas, C.; Pierotti, I.; Pompaiah, M.; Wallaschek, N.; Germer, C.T.; Wiegering, A.; Bartfeld, S. Location-specific cell identity rather than exposure to GI microbiota defines many innate immune signalling cascades in the gut epithelium. Gut 2021, 70, 687–697. [Google Scholar] [CrossRef]
- Ribatti, D. Tissue factor in hematological malignancies. Leukemia. 2006, 20, 1356–1357. [Google Scholar] [CrossRef] [Green Version]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [Green Version]
- Stockmann, C.; Schadendorf, D.; Klose, R.; Helfrich, I. The impact of the immune system on tumor: Angiogenesis and vascular remodeling. Front. Oncol. 2014, 4, 69. [Google Scholar] [CrossRef] [Green Version]
- McCourt, M.; Wang, J.H.; Sookhai, S.; Redmond, H.P. Proinflammatory mediators stimulate neutrophil-directed angiogenesis. Arch. Surg. 1999, 134, 1325–1331, discussion 1331-2. [Google Scholar] [CrossRef]
- Fritsch, M.; Günther, S.D.; Schwarzer, R.; Albert, M.C.; Schorn, F.; Werthenbach, J.P.; Schiffmann, L.M.; Stair, N.; Stocks, H.; Seeger, J.M.; et al. Caspase-8 is the molecular switch for apoptosis, necroptosis and pyroptosis. Nature 2019, 575, 683–687. [Google Scholar] [CrossRef]
- Ning, Y.; Manegold, P.C.; Hong, Y.K.; Zhang, W.; Pohl, A.; Lurje, G.; Winder, T.; Yang, D.; LaBonte, M.J.; Wilson, P.M.; et al. Interleukin-8 is associated with proliferation, migration, angiogenesis and chemosensitivity in vitro and in vivo in colon cancer cell line models. Int. J. Cancer 2011, 128, 2038–2049. [Google Scholar] [CrossRef] [Green Version]
- Bandapalli, O.R.; Ehrmann, F.; Ehemann, V.; Gaida, M.; Macher-Goeppinger, S.; Wente, M.; Schirmacher, P.; Brand, K. Down-regulation of CXCL1 inhibits tumor growth in colorectal liver metastasis. Cytokine 2012, 57, 46–53. [Google Scholar] [CrossRef]
- Sinnamon, M.J.; Carter, K.J.; Fingleton, B.; Matrisian, L.M. Matrix metalloproteinase-9 contributes to intestinal tumourigenesis in the adenomatous polyposis coli multiple intestinal neoplasia mouse. Int. J. Exp. Pathol. 2008, 89, 466–475. [Google Scholar] [CrossRef] [PubMed]
- Sommer, F.; Nookaew, I.; Sommer, N.; Fogelstrand, P.; Bäckhed, F. Site-specific programming of the host epithelial transcriptome by the gut microbiota. Genome Biol. 2015, 16, 62. [Google Scholar] [CrossRef] [Green Version]
- Koppel, N.; Balskus, E.P. Exploring and understanding the biochemical diversity of the human microbiota. Cell Chem. Biol. 2016, 23, 18–30. [Google Scholar] [CrossRef] [Green Version]
- Hyland, N.P.; Cryan, J.F. Microbe-host interactions: Influence of the gut microbiota on the enteric nervous system. Dev. Biol. 2016, 417, 182–187. [Google Scholar] [CrossRef] [PubMed]
- Roosen, L.; Boesmans, W.; Dondeyne, M.; Depoortere, I.; Tack, J.; Vanden Berghe, P. Specific hunger- and satiety-induced tuning of guinea pig enteric nerve activity. J. Physiol. 2012, 590, 4321–4333. [Google Scholar] [CrossRef] [PubMed]
- Johnson, L.R. Gastrointestinal hormones and their funcitons. Annu. Rev. Physiol. 1977, 39, 135–158. [Google Scholar] [CrossRef] [PubMed]
- Ellis, M.; Chambers, J.D.; Gwynne, R.M.; Bornstein, J.C. Serotonin and cholecystokinin mediate nutrient-induced segmentation in guinea pig small intestine. Am. J. Physiol. Gastrointest. Liver Physiol. 2013, 304, G749–G761. [Google Scholar] [CrossRef] [Green Version]
- Dass, N.B.; Munonyara, M.; Bassil, A.K.; Hervieu, G.J.; Osbourne, S.; Corcoran, S.; Morgan, M.; Sanger, G.J. Growth hormone secretagogue receptors in rat and human gastrointestinal tract and the effects of ghrelin. Neuroscience 2003, 120, 443–453. [Google Scholar] [CrossRef]
- Waise, T.M.Z.; Dranse, H.J.; Lam, T.K.T. The metabolic role of vagal afferent innervation. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 625–636. [Google Scholar] [CrossRef]
- Duca, F.A.; Waise, T.M.Z.; Peppler, W.T.; Lam, T.K.T. The metabolic impact of small intestinal nutrient sensing. Nat. Commun. 2021, 12, 903. [Google Scholar] [CrossRef]
- Furness, J.B.; Rivera, L.R.; Cho, H.J.; Bravo, D.M.; Callaghan, B. The gut as a sensory organ. Nat. Rev. Gastroenterol. Hepatol. 2013, 10, 729–740. [Google Scholar] [CrossRef] [PubMed]
- Neunlist, M.; Schemann, M. Nutrient-induced changes in the phenotype and function of the enteric nervous system. J. Physiol. 2014, 592, 2959–2965. [Google Scholar] [CrossRef] [PubMed]
- Donia, M.S.; Fischbach, M.A. Human microbiota. Small molecules from the human microbiota. Science 2015, 349, 1254766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rolls, A.; Shechter, R.; London, A.; Ziv, Y.; Ronen, A.; Levy, R.; Schwartz, M. Toll-like receptors modulate adult hippocampal neurogenesis. Nat. Cell Biol. 2007, 9, 1081–1088. [Google Scholar] [CrossRef] [PubMed]
- Brun, P.; Giron, M.C.; Qesari, M.; Porzionato, A.; Caputi, V.; Zoppellaro, C.; Banzato, S.; Grillo, A.R.; Spagnol, L.; De Caro, R.; et al. Toll-like receptor 2 regulates intestinal inflammation by controlling integrity of the enteric nervous system. Gastroenterology 2013, 145, 1323–1333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bogunovic, M.; Ginhoux, F.; Helft, J.; Shang, L.; Hashimoto, D.; Greter, M.; Liu, K.; Jakubzick, C.; Ingersoll, M.A.; Leboeuf, M.; et al. Origin of the lamina propria dendritic cell network. Immunity 2009, 31, 513–525. [Google Scholar] [CrossRef] [Green Version]
- Schemann, M.; Camilleri, M. Functions and imaging of mast cell and neural axis of the gut. Gastroenterology 2013, 144, 698–704.e4. [Google Scholar] [CrossRef] [Green Version]
- Muller, P.A.; Koscsó, B.; Rajani, G.M.; Stevanovic, K.; Berres, M.L.; Hashimoto, D.; Mortha, A.; Leboeuf, M.; Li, X.M.; Mucida, D.; et al. Crosstalk between muscularis macrophages and enteric neurons regulates gastrointestinal motility. Cell 2014, 158, 300–313. [Google Scholar] [CrossRef] [Green Version]
- Avetisyan, M.; Rood, J.E.; Huerta Lopez, S.; Sengupta, R.; Wright-Jin, E.; Dougherty, J.D.; Behrens, E.M.; Heuckeroth, R.O. Muscularis macrophage development in the absence of an enteric nervous system. Proc. Natl. Acad. Sci. USA 2018, 115, 4696–4701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gabanyi, I.; Muller, P.A.; Feighery, L.; Oliveira, T.Y.; Costa-Pinto, F.A.; Mucida, D. Neuro-immune interactions drive tissue programming in intestinal macrophages. Cell 2016, 164, 378–391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matheis, F.; Muller, P.A.; Graves, C.L.; Gabanyi, I.; Kerner, Z.J.; Costa-Borges, D.; Ahrends, T.; Rosenstiel, P.; Mucida, D. Adrenergic signaling in muscularis macrophages limits infection-induced neuronal loss. Cell 2020, 180, 64–78. [Google Scholar] [CrossRef]
- Van Diest, S.A.; Stanisor, O.I.; Boeckxstaens, G.E.; de Jonge, W.J.; van den Wijngaard, R.M. Relevance of mast cell-nerve interactions in intestinal nociception. Biochim. Biophys. Acta 2012, 1822, 74–84. [Google Scholar] [CrossRef] [Green Version]
- Jacobson, A.; Yang, D.; Vella, M.; Chiu, I.M. The intestinal neuro-immune axis: Crosstalk between neurons, immune cells, and microbes. Mucosal Immunol. 2021, 14, 555–565. [Google Scholar] [CrossRef]
- Godinho-Silva, C.; Cardoso, F.; Veiga-Fernandes, H. Neuro-immune cell units: A new paradigm in physiology. Annu. Rev. Immunol. 2019, 37, 19–46. [Google Scholar] [CrossRef] [Green Version]
- Veiga-Fernandes, H.; Mucida, D. Neuro-immune interactions at barrier surfaces. Cell 2016, 165, 801–811. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Collins, J.; Borojevic, R.; Verdu, E.F.; Huizinga, J.D.; Ratcliffe, E.M. Intestinal microbiota influence the early postnatal development of the enteric nervous system. Neurogastroenterol. Motil. 2014, 26, 98–107. [Google Scholar] [CrossRef]
- De Vadder, F.; Grasset, E.; Mannerås Holm, L.; Karsenty, G.; Macpherson, A.J.; Olofsson, L.E.; Bäckhed, F. Gut microbiota regulates maturation of the adult enteric nervous system via enteric serotonin networks. Proc. Natl. Acad. Sci. USA 2018, 115, 6458–6463. [Google Scholar] [CrossRef] [Green Version]
- Heiss, C.N.; Olofsson, L.E. The role of the gut microbiota in development, function and disorders of the central nervous system and the enteric nervous system. J. Neuroendocrinol. 2019, 31, e12684. [Google Scholar] [CrossRef]
- Lomasney, K.W.; Houston, A.; Shanahan, F.; Dinan, T.G.; Cryan, J.F.; Hyland, N.P. Selective influence of host microbiota on cAMP-mediated ion transport in mouse colon. Neurogastroenterol. Motil. 2014, 26, 887–890. [Google Scholar] [CrossRef]
- McVey Neufeld, K.A.; Mao, Y.K.; Bienenstock, J.; Foster, J.A.; Kunze, W.A. The microbiome is essential for normal gut intrinsic primary afferent neuron excitability in the mouse. Neurogastroenterol. Motil. 2013, 25, 183–190. [Google Scholar] [CrossRef]
- Aktar, R.; Parkar, N.; Stentz, R.; Baumard, L.; Parker, A.; Goldson, A.; Brion, A.; Carding, S.; Blackshaw, A.; Peiris, M. Human resident gut microbe Bacteroides thetaiotaomicron regulates colonic neuronal innervation and neurogenic function. Gut Microbes 2020, 11, 1745–1757. [Google Scholar] [CrossRef] [PubMed]
- Olsson, C.; Holmgren, S. The control of gut motility. Comp. Biochem. Physiol. A. Mol. Integr. Physiol. 2001, 128, 481–503. [Google Scholar] [CrossRef]
- Jacobs, L.R. Differential effects of dietary fibers on rat intestinal circular muscle cell size. Dig. Dis. Sci. 1985, 30, 247–252. [Google Scholar] [CrossRef] [PubMed]
- Le Blay, G.; Blottière, H.M.; Ferrier, L.; Le Foll, E.; Bonnet, C.; Galmiche, J.P.; Cherbut, C. Short-chain fatty acids induce cytoskeletal and extracellular protein modifications associated with modulation of proliferation on primary culture of rat intestinal smooth muscle cells. Dig. Dis. Sci. 2000, 45, 1623–1630. [Google Scholar] [CrossRef]
- Bartholome, A.L.; Albin, D.M.; Baker, D.H.; Holst, J.J.; Tappenden, K.A. Supplementation of total parenteral nutrition with butyrate acutely increases structural aspects of intestinal adaptation after an 80% jejunoileal resection in neonatal piglets. JPEN J. Parenter. Enteral Nutr. 2004, 28, 210–222, discussion 222–223. [Google Scholar] [CrossRef]
- Mitsui, R.; Karaki, S.I.; Kubo, Y.; Sugiura, Y.; Kuwahara, A. Fibre-free diet leads to impairment of neuronally mediated muscle contractile response in rat distal colon. Neurogastroenterol. Motil. 2006, 18, 1093–1101. [Google Scholar] [CrossRef]
- Meritt, J.; Witkowski, T.A.; Nagele, R.; Norcross, E.D.; Stein, T.P. Glutamine and smooth muscle morphology of the gut in rats on total parenteral nutrition. J. Am. Coll. Nutr. 1989, 8, 537–544. [Google Scholar] [CrossRef]
- Zhu, J.; Wu, Y.; Guo, Y.; Tang, Q.; Lu, T.; Cai, W.; Huang, H. Choline alleviates parenteral nutrition-associated duodenal motility disorder in infant rats. JPEN J. Parenter. Enteral Nutr. 2016, 40, 995–1005. [Google Scholar] [CrossRef]
- Vrabcova, M.; Mikuska, L.; Vazan, R.; Miko, M.; Varga, I.; Mravec, B. Effect of chronic intake of liquid nutrition on stomach and duodenum morphology. Acta Histochem. 2016, 118, 435–442. [Google Scholar] [CrossRef]
- Phillips, R.J.; Powley, T.L. Macrophages associated with the intrinsic and extrinsic autonomic innervation of the rat gastrointestinal tract. Auton. Neurosci. 2012, 169, 12–27. [Google Scholar] [CrossRef] [Green Version]
- Ji, S.; Traini, C.; Mischopoulou, M.; Gibbons, S.J.; Ligresti, G.; Faussone-Pellegrini, M.S.; Sha, L.; Farrugia, G.; Vannucchi, M.G.; Cipriani, G. Muscularis macrophages establish cell-to-cell contacts with telocytes/PDGFRα-positive cells and smooth muscle cells in the human and mouse gastrointestinal tract. Neurogastroenterol. Motil. 2021, 33, e13993. [Google Scholar] [CrossRef]
- Viola, M.F.; Boeckxstaens, G. Intestinal resident macrophages: Multitaskers of the gut. Neurogastroenterol. Motil. 2020, 32, e13843. [Google Scholar] [CrossRef] [Green Version]
- Nair, D.G.; Miller, K.G.; Lourenssen, S.R.; Blennerhassett, M.G. Inflammatory cytokines promote growth of intestinal smooth muscle cells by induced expression of PDGF-Rβ. J. Cell. Mol. Med. 2014, 18, 444–454. [Google Scholar] [CrossRef]
- Blennerhassett, M.G.; Bovell, F.M.; Lourenssen, S.; McHugh, K.M. Characteristics of inflammation-induced hypertrophy of rat intestinal smooth muscle cell. Dig. Dis. Sci. 1999, 44, 1265–1272. [Google Scholar] [CrossRef]
- Guarino, M.P.; Cicala, M.; Putignani, L.; Severi, C. Gastrointestinal neuromuscular apparatus: An underestimated target of gut microbiota. World J. Gastroenterol. 2016, 22, 9871–9879. [Google Scholar] [CrossRef]
- Brun, P.; Gobbo, S.; Caputi, V.; Spagnol, L.; Schirato, G.; Pasqualin, M.; Levorato, E.; Palù, G.; Giron, M.C.; Castagliuolo, I. Toll like receptor-2 regulates production of glial-derived neurotrophic factors in murine intestinal smooth muscle cells. Mol. Cell. Neurosci. 2015, 68, 24–35. [Google Scholar] [CrossRef]
- Kamath, P.S.; Phillips, S.F.; Zinsmeister, A.R. Short-chain fatty acids stimulate ileal motility in humans. Gastroenterology 1988, 95, 1496–1502. [Google Scholar] [CrossRef]
- Squires, P.E.; Rumsey, R.D.; Edwards, C.A.; Read, N.W. Effect of short-chain fatty acids on contractile activity and fluid flow in rat colon in vitro. Am. J. Physiol. 1992, 262 Pt 1, G813–G817. [Google Scholar] [CrossRef]
- Yarullina, D.R.; Shafigullin, M.U.; Sakulin, K.A.; Arzamastseva, A.A.; Shaidullov, I.F.; Markelova, M.I.; Grigoryeva, T.V.; Karpukhin, O.Y.; Sitdikova, G.F. Characterization of gut contractility and microbiota in patients with severe chronic constipation. PLoS ONE 2020, 15, e0235985. [Google Scholar] [CrossRef]
- Gibson, G.R.; Hutkins, R.; Sanders, M.E.; Prescott, S.L.; Reimer, R.A.; Salminen, S.J.; Scott, K.; Stanton, C.; Swanson, K.S.; Cani, P.D.; et al. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 491–502. [Google Scholar] [CrossRef] [Green Version]
- Murga-Garrido, S.M.; Hong, Q.; Cross, T.-W.L.; Hutchison, E.R.; Han, J.; Thomas, S.P.; Vivas, E.I.; Denu, J.; Ceschin, D.G.; Tang, Z.-Z.; et al. Gut microbiome variation modulates the effects of dietary fiber on host metabolism. Microbiome 2021, 9, 117. [Google Scholar] [CrossRef]
- Man, A.W.C.; Zhou, Y.; Xia, N.; Li, H. Involvement of gut microbiota, microbial metabolites and interaction with polyphenol in host immunometabolism. Nutrients 2020, 12, 3054. [Google Scholar] [CrossRef]
- Muccioli, G.G.; Naslain, D.; Bäckhed, F.; Reigstad, C.S.; Lambert, D.M.; Delzenne, N.M.; Cani, P.D. The endocannabinoid system links gut microbiota to adipogenesis. Mol. Syst. Biol. 2010, 6, 392. [Google Scholar] [CrossRef]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bayer, F.; Dremova, O.; Khuu, M.P.; Mammadova, K.; Pontarollo, G.; Kiouptsi, K.; Soshnikova, N.; May-Simera, H.L.; Endres, K.; Reinhardt, C. The Interplay between Nutrition, Innate Immunity, and the Commensal Microbiota in Adaptive Intestinal Morphogenesis. Nutrients 2021, 13, 2198. https://0-doi-org.brum.beds.ac.uk/10.3390/nu13072198
Bayer F, Dremova O, Khuu MP, Mammadova K, Pontarollo G, Kiouptsi K, Soshnikova N, May-Simera HL, Endres K, Reinhardt C. The Interplay between Nutrition, Innate Immunity, and the Commensal Microbiota in Adaptive Intestinal Morphogenesis. Nutrients. 2021; 13(7):2198. https://0-doi-org.brum.beds.ac.uk/10.3390/nu13072198
Chicago/Turabian StyleBayer, Franziska, Olga Dremova, My Phung Khuu, Könül Mammadova, Giulia Pontarollo, Klytaimnistra Kiouptsi, Natalia Soshnikova, Helen Louise May-Simera, Kristina Endres, and Christoph Reinhardt. 2021. "The Interplay between Nutrition, Innate Immunity, and the Commensal Microbiota in Adaptive Intestinal Morphogenesis" Nutrients 13, no. 7: 2198. https://0-doi-org.brum.beds.ac.uk/10.3390/nu13072198