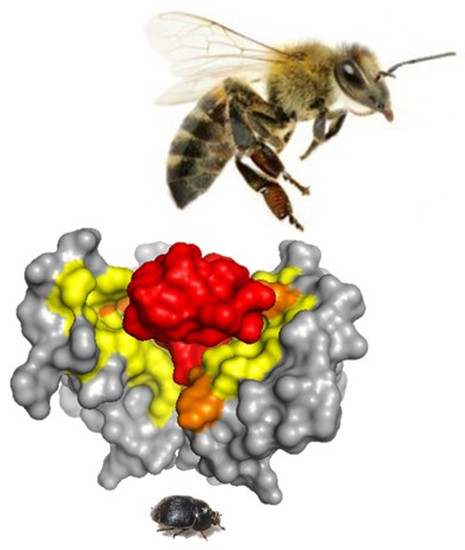
Identification of Aethina tumida Kir Channels as Putative Targets of the Bee Venom Peptide Tertiapin Using Structure-Based Virtual Screening Methods
Abstract
:1. Introduction
2. Results
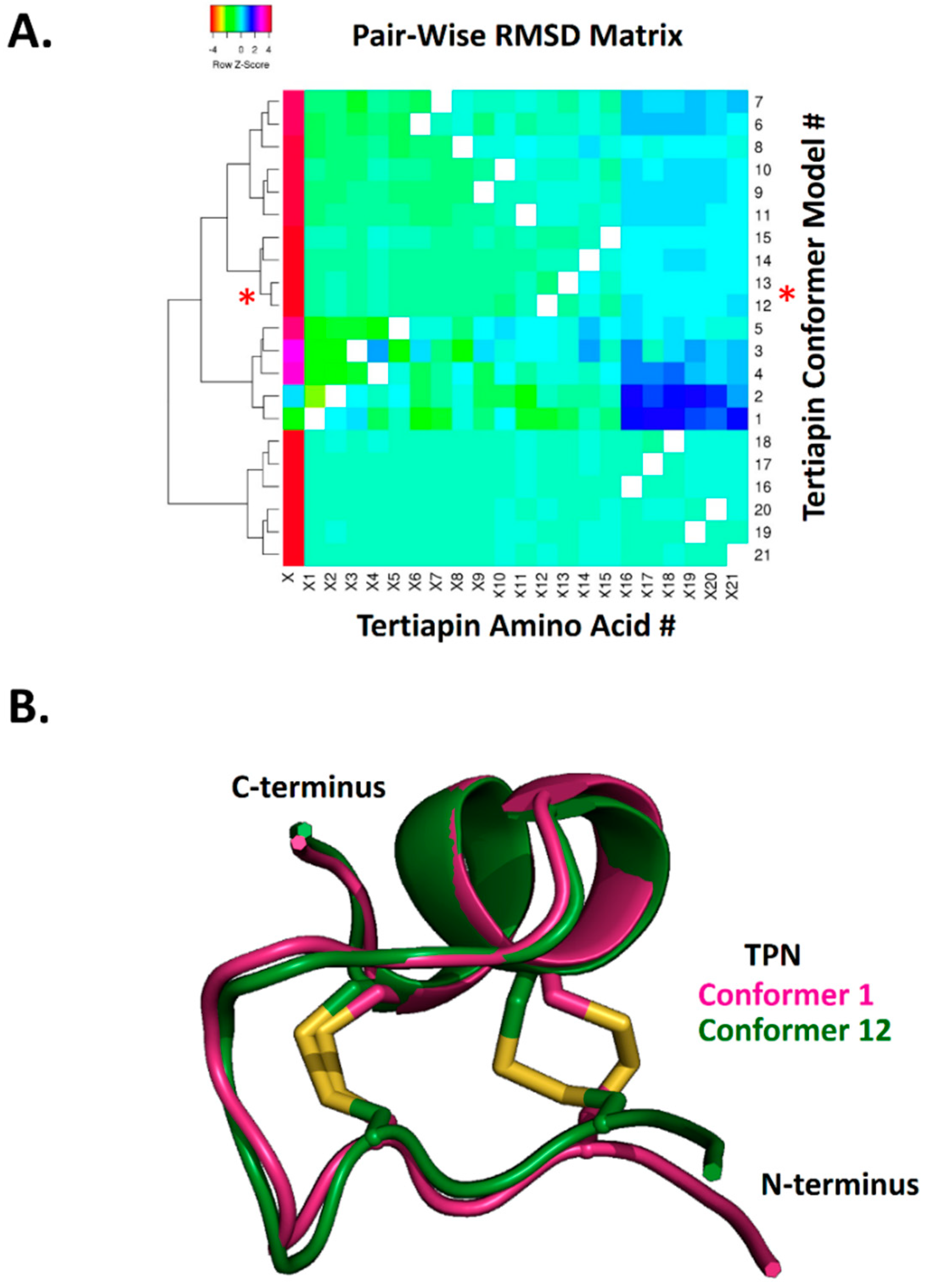
2.1. Reliability of Molecular Docking of TPN to Kir Channel-Modelled Structures
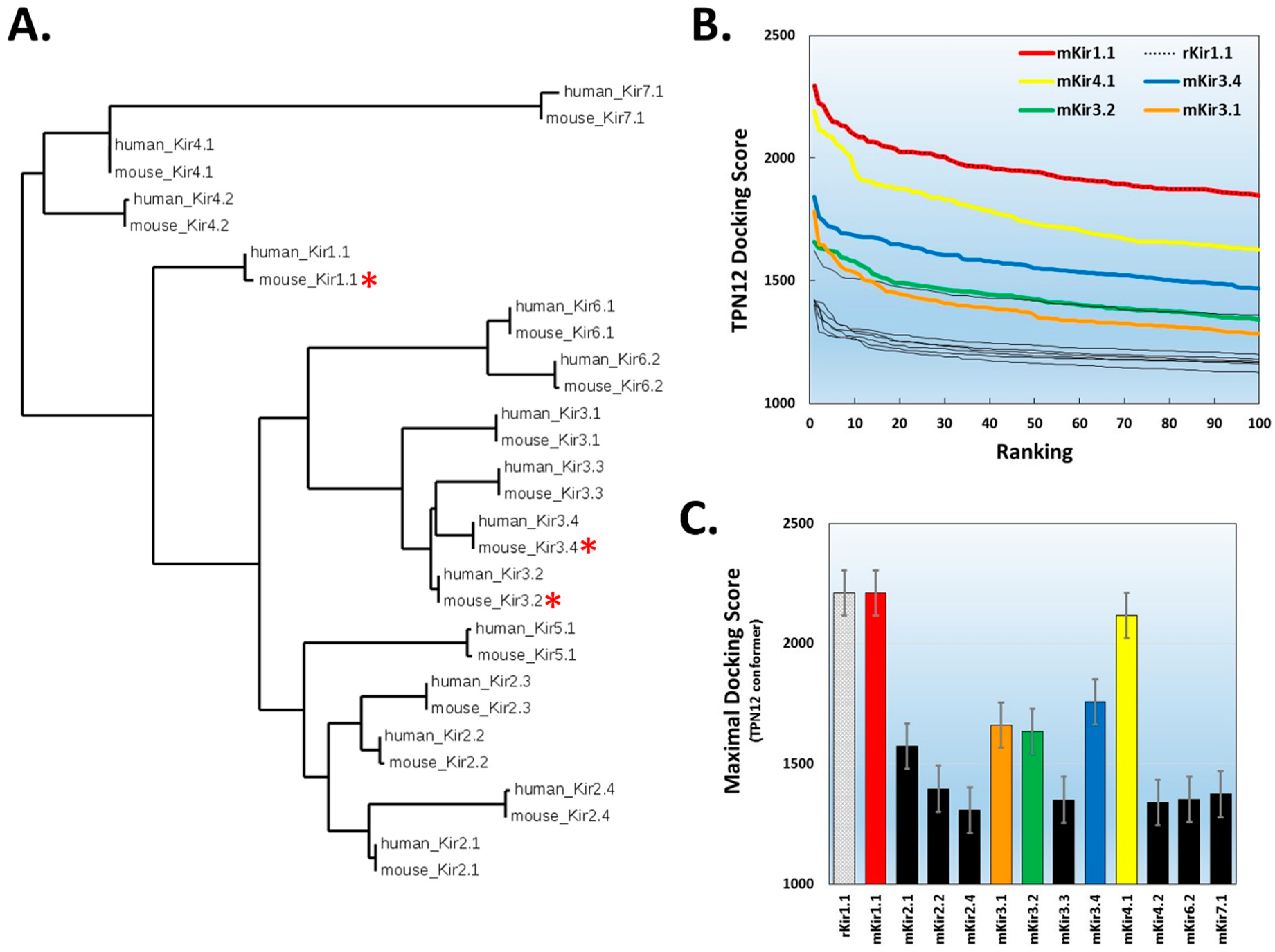
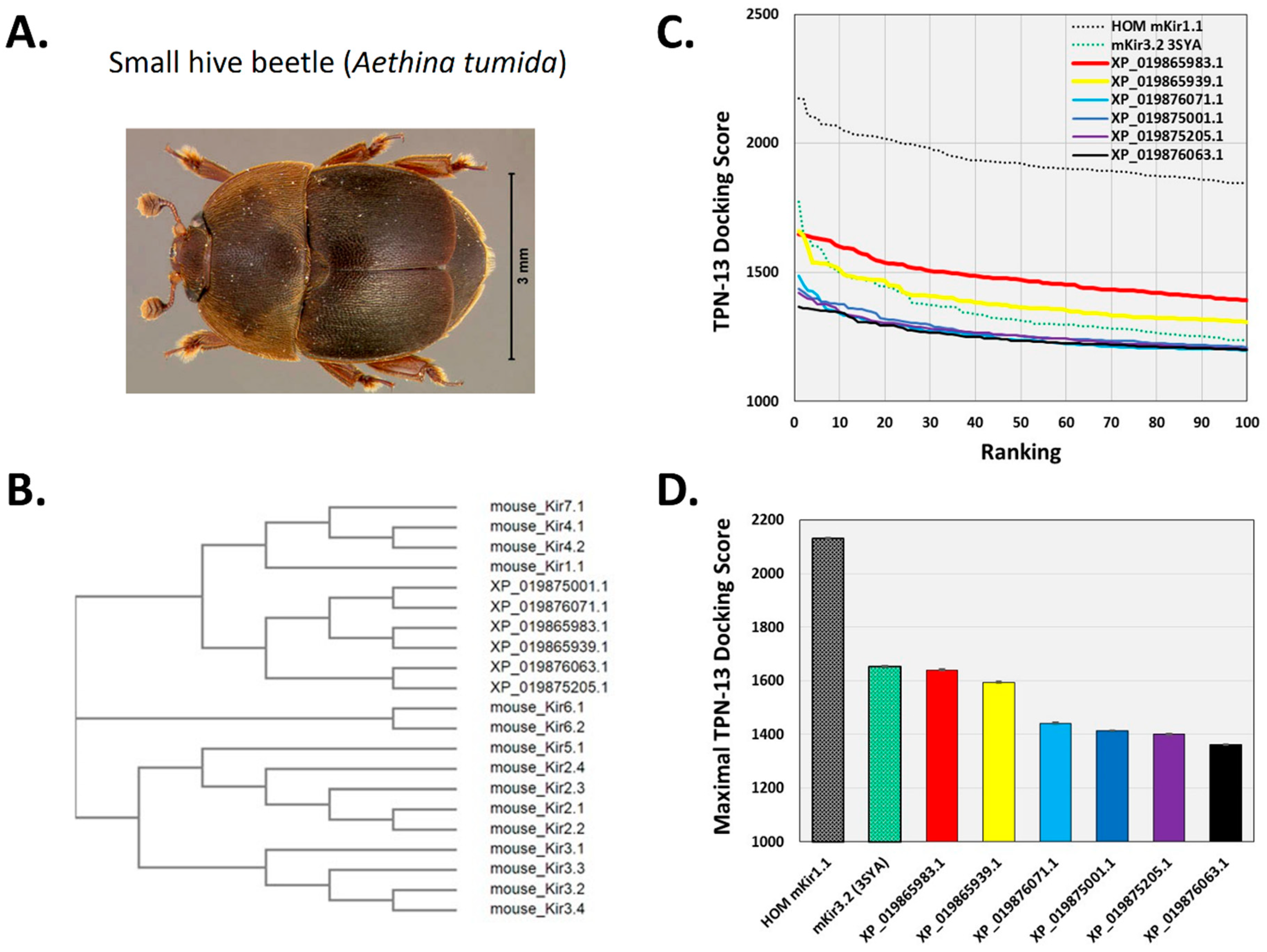
2.2. Virtual Screening for TPN-Interacting Kir Channels
2.3. Interface Analysis of TPN-Docked Kir Channels
2.4. Testing TPNQ Block of Kir4.1 Channels Expressed in Xenopus Oocytes
2.5. Hypothesis Testing with Structure-Based Virtual Screening
3. Discussion
4. Materials and Methods
4.1. Ion Channel Structures and Homology Modelling
4.2. Computational Docking of TPN to Kir Channel Structures
4.3. Kir Channel–TPN Interface Analysis
4.4. Kir Channel Expression in Xenopus Oocytes
4.5. Two-Electrode Voltage Clamp Recording from Xenopus Oocytes
Funding
Conflicts of Interest
References
- Daly, N.L.; Wilson, D. Structural diversity of arthropod venom toxins. Toxicon Off. J. Int. Soc. Toxinol. 2018, 152, 46–56. [Google Scholar] [CrossRef] [PubMed]
- de Graaf, D.C.; Aerts, M.; Danneels, E.; Devreese, B. Bee, wasp and ant venomics pave the way for a component-resolved diagnosis of sting allergy. J. Proteom. 2009, 72, 145–154. [Google Scholar] [CrossRef] [PubMed]
- Casewell, N.R.; Wuster, W.; Vonk, F.J.; Harrison, R.A.; Fry, B.G. Complex cocktails: The evolutionary novelty of venoms. Trends Ecol. Evol. 2013, 28, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Fry, B.G.; Roelants, K.; Champagne, D.E.; Scheib, H.; Tyndall, J.D.; King, G.F.; Nevalainen, T.J.; Norman, J.A.; Lewis, R.J.; Norton, R.S.; et al. The toxicogenomic multiverse: Convergent recruitment of proteins into animal venoms. Annu. Rev. Genom. Hum. Genet. 2009, 10, 483–511. [Google Scholar] [CrossRef] [PubMed]
- Kalia, J.; Milescu, M.; Salvatierra, J.; Wagner, J.; Klint, J.K.; King, G.F.; Olivera, B.M.; Bosmans, F. From foe to friend: Using animal toxins to investigate ion channel function. J. Mol. Biol. 2015, 427, 158–175. [Google Scholar] [CrossRef] [PubMed]
- Gordon, D.; Chen, R.; Chung, S.H. Computational methods of studying the binding of toxins from venomous animals to biological ion channels: Theory and applications. Physiol. Rev. 2013, 93, 767–802. [Google Scholar] [CrossRef] [PubMed]
- Doupnik, C.A.; Parra, K.C.; Guida, W.C. A computational design approach for virtual screening of peptide interactions across K(+) channel families. Comput. Struct. Biotechnol. J. 2015, 13, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Rashid, M.H.; Huq, R.; Tanner, M.R.; Chhabra, S.; Khoo, K.K.; Estrada, R.; Dhawan, V.; Chauhan, S.; Pennington, M.W.; Beeton, C.; et al. A potent and Kv1.3-selective analogue of the scorpion toxin HsTX1 as a potential therapeutic for autoimmune diseases. Sci. Rep. 2014, 4, 4509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pennington, M.W.; Harunur Rashid, M.; Tajhya, R.B.; Beeton, C.; Kuyucak, S.; Norton, R.S. A C-terminally amidated analogue of ShK is a potent and selective blocker of the voltage-gated potassium channel Kv1.3. FEBS Lett. 2012, 586, 3996–4001. [Google Scholar] [CrossRef] [Green Version]
- Lewis, R.J.; Garcia, M.L. Therapeutic potential of venom peptides. Nat. Rev. Drug Discov. 2003, 2, 790–802. [Google Scholar] [CrossRef]
- Elsik, C.G.; Worley, K.C.; Bennett, A.K.; Beye, M.; Camara, F.; Childers, C.P.; de Graaf, D.C.; Debyser, G.; Deng, J.; Devreese, B.; et al. Finding the missing honey bee genes: Lessons learned from a genome upgrade. BMC Genom. 2014, 15, 86. [Google Scholar] [CrossRef] [PubMed]
- Gauldie, J.; Hanson, J.M.; Rumjanek, F.D.; Shipolini, R.A.; Vernon, C.A. The peptide components of bee venom. Eur. J. Biochem. 1976, 61, 369–376. [Google Scholar] [CrossRef] [PubMed]
- Doupnik, C.A. Venom-derived peptides inhibiting Kir channels: Past, present, and future. Neuropharmacology 2017, 127, 161–172. [Google Scholar] [CrossRef] [PubMed]
- Jin, W.; Lu, Z. A novel high-affinity inhibitor for inward-rectifier K+ channels. Biochemistry 1998, 37, 13291–13299. [Google Scholar] [CrossRef] [PubMed]
- Felix, J.P.; Liu, J.; Schmalhofer, W.A.; Bailey, T.; Bednarek, M.A.; Kinkel, S.; Weinglass, A.B.; Kohler, M.; Kaczorowski, G.J.; Priest, B.T.; et al. Characterization of Kir1.1 channels with the use of a radiolabeled derivative of tertiapin. Biochemistry 2006, 45, 10129–10139. [Google Scholar] [CrossRef] [PubMed]
- Jin, W.; Klem, A.M.; Lewis, J.H.; Lu, Z. Mechanisms of inward-rectifier K+ channel inhibition by tertiapin-Q. Biochemistry 1999, 38, 14294–14301. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Nelson, J.W. Solution structure of tertiapin determined using nuclear magnetic resonance and distance geometry. Proteins 1993, 17, 124–137. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Qiu, S.; Yang, F.; Cao, Z.; Li, W.; Wu, Y. Unique mechanism of the interaction between honey bee toxin TPNQ and rKir1.1 potassium channel explored by computational simulations: Insights into the relative insensitivity of channel towards animal toxins. PLoS ONE 2013, 8, e67213. [Google Scholar] [CrossRef] [PubMed]
- Whorton, M.R.; MacKinnon, R. X-ray structure of the mammalian GIRK2-betagamma G-protein complex. Nature 2013, 498, 190–197. [Google Scholar] [CrossRef]
- Whorton, M.R.; MacKinnon, R. Crystal structure of the mammalian GIRK2 K+ channel and gating regulation by G proteins, PIP2, and sodium. Cell 2011, 147, 199–208. [Google Scholar] [CrossRef]
- Hibino, H.; Inanobe, A.; Furutani, K.; Murakami, S.; Findlay, I.; Kurachi, Y. Inwardly rectifying potassium channels: Their structure, function, and physiological roles. Physiol. Rev. 2010, 90, 291–366. [Google Scholar] [CrossRef] [PubMed]
- Lesage, F.; Guillemare, E.; Fink, M.; Duprat, F.; Heurteaux, C.; Fosset, M.; Romey, G.; Barhanin, J.; Lazdunski, M. Molecular properties of neuronal G-protein-activated inwardly rectifying K+ channels. J. Biol. Chem. 1995, 270, 28660–28667. [Google Scholar] [CrossRef] [PubMed]
- Duprat, F.; Lesage, F.; Guillemare, E.; Fink, M.; Hugnot, J.P.; Bigay, J.; Lazdunski, M.; Romey, G.; Barhanin, J. Heterologous multimeric assembly is essential for K+ channel activity of neuronal and cardiac G-protein-activated inward rectifiers. Biochem. Biophys. Res. Commun. 1995, 212, 657–663. [Google Scholar] [CrossRef] [PubMed]
- Kofuji, P.; Davidson, N.; Lester, H.A. Evidence that neuronal G-protein-gated inwardly rectifying K+ channels are activated by G beta gamma subunits and function as heteromultimers. Proc. Natl. Acad. Sci. USA 1995, 92, 6542–6546. [Google Scholar] [CrossRef] [PubMed]
- Pierce, B.G.; Wiehe, K.; Hwang, H.; Kim, B.H.; Vreven, T.; Weng, Z. ZDOCK server: Interactive docking prediction of protein-protein complexes and symmetric multimers. Bioinformatics 2014, 30, 1771–1773. [Google Scholar] [CrossRef] [PubMed]
- Ramu, Y.; Klem, A.M.; Lu, Z. Short variable sequence acquired in evolution enables selective inhibition of various inward-rectifier K+ channels. Biochemistry 2004, 43, 10701–10709. [Google Scholar] [CrossRef] [PubMed]
- Ramu, Y.; Xu, Y.; Lu, Z. Engineered specific and high-affinity inhibitor for a subtype of inward-rectifier K+ channels. Proc. Natl. Acad. Sci. USA 2008, 105, 10774–10778. [Google Scholar] [CrossRef]
- Krissinel, E.; Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007, 372, 774–797. [Google Scholar] [CrossRef]
- Santos, L.D.; Pieroni, M.; Menegasso, A.R.S.; Pinto, J.R.A.S.; Palma, M.S. A new scenario of bioprospecting of Hymenoptera venoms through proteomic approach. J. Venom. Anim. Toxins 2011, 17, 364–377. [Google Scholar]
- Evans, J.D.; McKenna, D.; Scully, E.; Cook, S.C.; Dainat, B.; Egekwu, N.; Grubbs, N.; Lopez, D.; Lorenzen, M.D.; Reyna, S.M.; et al. Genome of the small hive beetle (Aethina tumida, Coleoptera: Nitidulidae), a worldwide parasite of social bee colonies, provides insights into detoxification and herbivory. GigaScience 2018, 7, giy138. [Google Scholar] [CrossRef]
- Tarver, M.R.; Huang, Q.; de Guzman, L.; Rinderer, T.; Holloway, B.; Reese, J.; Weaver, D.; Evans, J.D. Transcriptomic and functional resources for the small hive beetle Aethina tumida, a worldwide parasite of honey bees. Genom. Data 2016, 9, 97–99. [Google Scholar] [CrossRef] [PubMed]
- Neumann, P.; Pettis, J.S.; Schazfer, M.O. Quo vadis Aethina tumida? Biology and control of small hive beetles. Apidologie 2016, 47, 427–466. [Google Scholar] [CrossRef] [Green Version]
- Nene, V.; Wortman, J.R.; Lawson, D.; Haas, B.; Kodira, C.; Tu, Z.J.; Loftus, B.; Xi, Z.; Megy, K.; Grabherr, M.; et al. Genome sequence of Aedes aegypti, a major arbovirus vector. Science 2007, 316, 1718–1723. [Google Scholar] [CrossRef] [PubMed]
- Luan, Z.; Li, H.S. Inwardly rectifying potassium channels in Drosophila. Sheng Li Xue Bao [Acta Physiol. Sin.] 2012, 64, 515–519. [Google Scholar] [PubMed]
- Beyenbach, K.W.; Yu, Y.; Piermarini, P.M.; Denton, J. Targeting renal epithelial channels for the control of insect vectors. Tissue Barriers 2015, 3, e1081861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geng, C.; Narasimhan, S.; Rodrigues, J.P.; Bonvin, A.M. Information-Driven, Ensemble Flexible Peptide Docking Using HADDOCK. Methods Mol. Biol. 2017, 1561, 109–138. [Google Scholar] [CrossRef] [Green Version]
- Mintseris, J.; Pierce, B.; Wiehe, K.; Anderson, R.; Chen, R.; Weng, Z. Integrating statistical pair potentials into protein complex prediction. Proteins 2007, 69, 511–520. [Google Scholar] [CrossRef]
- Comeau, S.R.; Gatchell, D.W.; Vajda, S.; Camacho, C.J. ClusPro: A fully automated algorithm for protein-protein docking. Nucleic Acids Res. 2004, 32, W96–W99. [Google Scholar] [CrossRef]
- Krissinel, E. Crystal contacts as nature’s docking solutions. J. Comput. Chem. 2010, 31, 133–143. [Google Scholar] [CrossRef]
- Doupnik, C.A.; Jaen, C.; Zhang, Q. Measuring the modulatory effects of RGS proteins on GIRK channels. Methods Enzymol. 2004, 389, 131–154. [Google Scholar] [CrossRef]







© 2019 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Doupnik, C.A. Identification of Aethina tumida Kir Channels as Putative Targets of the Bee Venom Peptide Tertiapin Using Structure-Based Virtual Screening Methods. Toxins 2019, 11, 546. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins11090546
Doupnik CA. Identification of Aethina tumida Kir Channels as Putative Targets of the Bee Venom Peptide Tertiapin Using Structure-Based Virtual Screening Methods. Toxins. 2019; 11(9):546. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins11090546
Chicago/Turabian StyleDoupnik, Craig A. 2019. "Identification of Aethina tumida Kir Channels as Putative Targets of the Bee Venom Peptide Tertiapin Using Structure-Based Virtual Screening Methods" Toxins 11, no. 9: 546. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins11090546