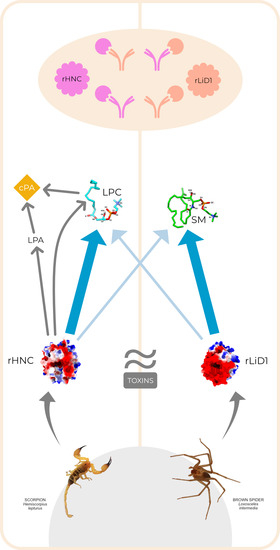
Antigenic and Substrate Preference Differences between Scorpion and Spider Dermonecrotic Toxins, a Comparative Investigation
Abstract
:1. Introduction
2. Results
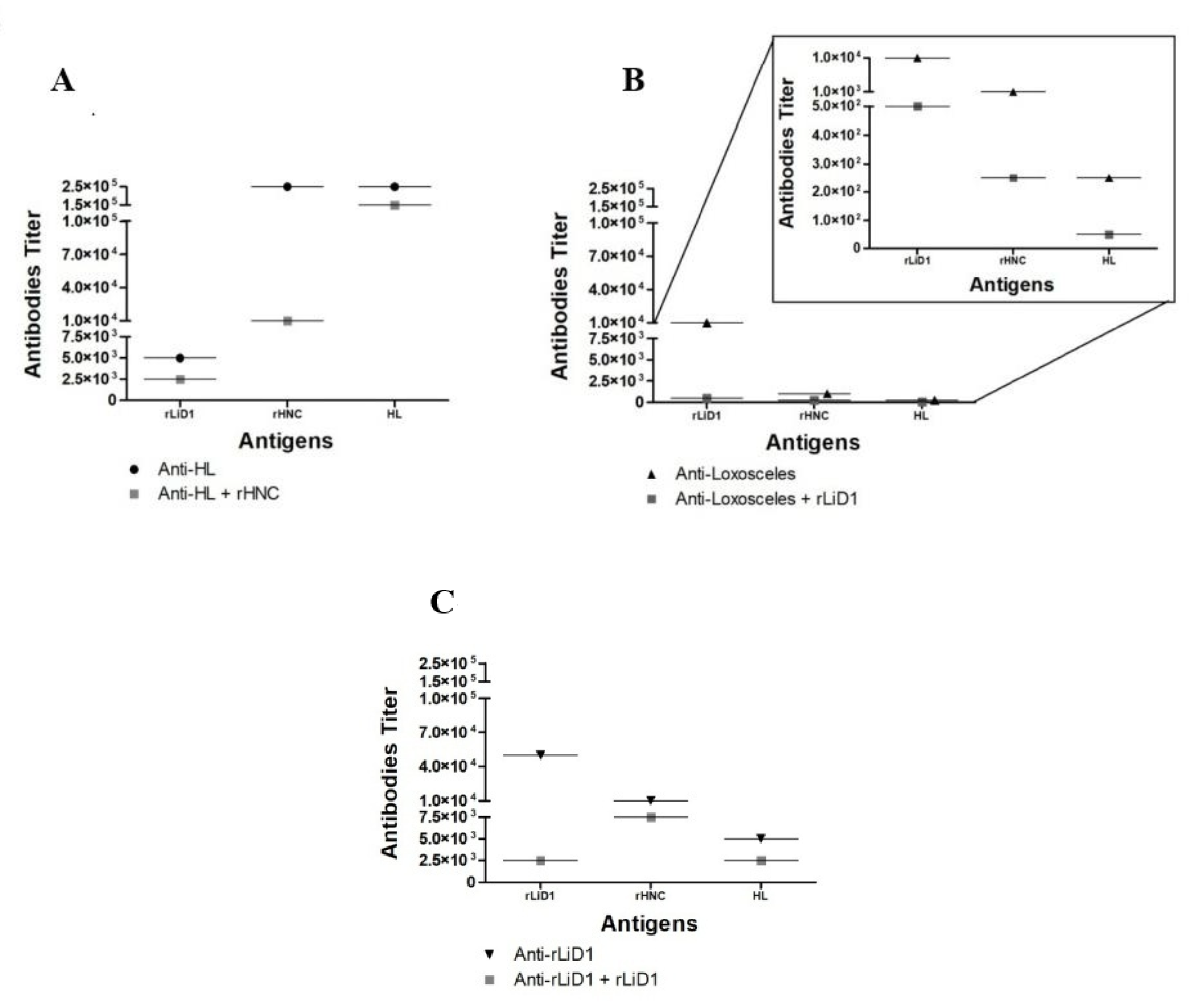
2.1. Cross-Reactivity of Anti-H. Lepturus and Anti-Loxosceles against rHNC and rLiD1
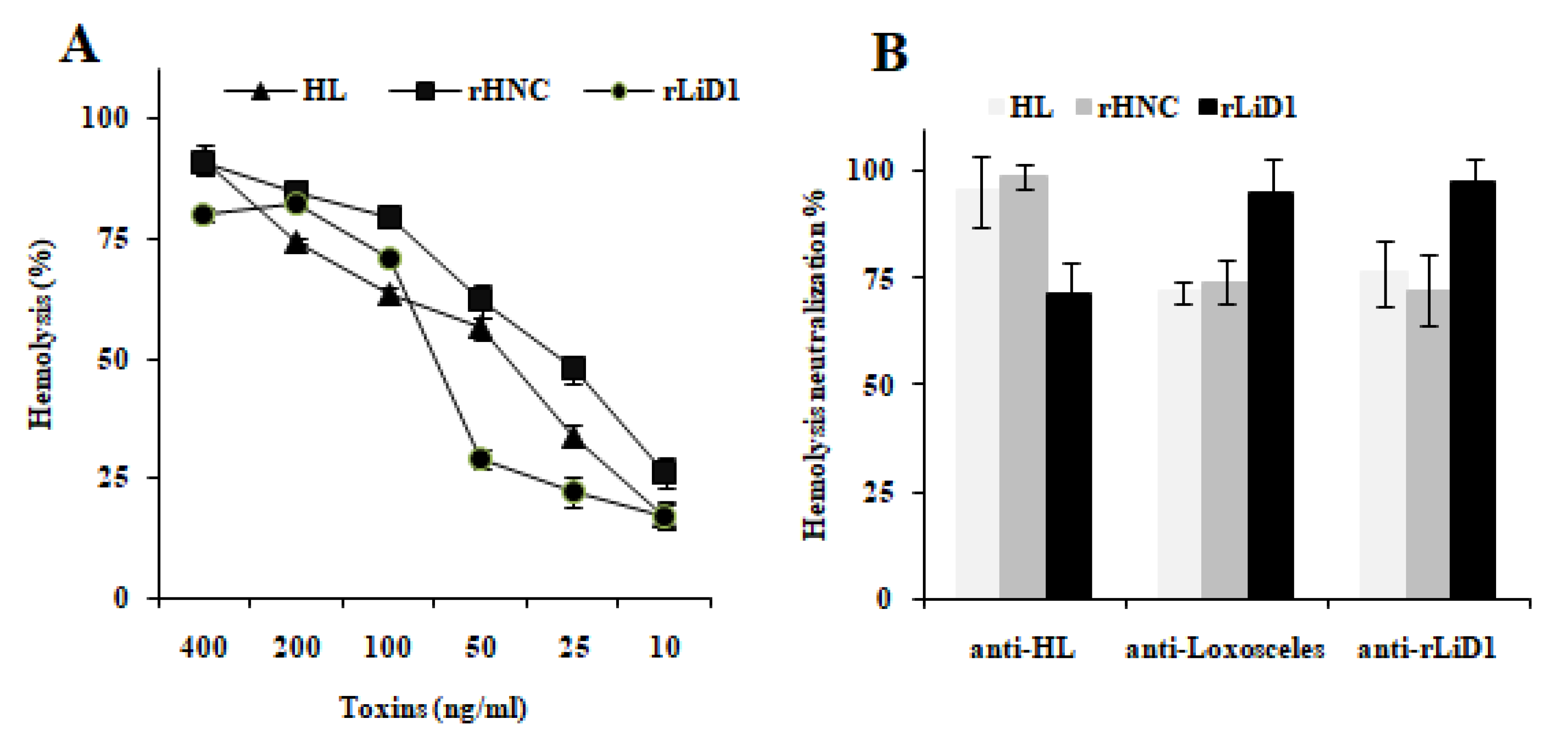
2.2. Cross-Neutralization of Complement-Dependent Hemolytic Activity
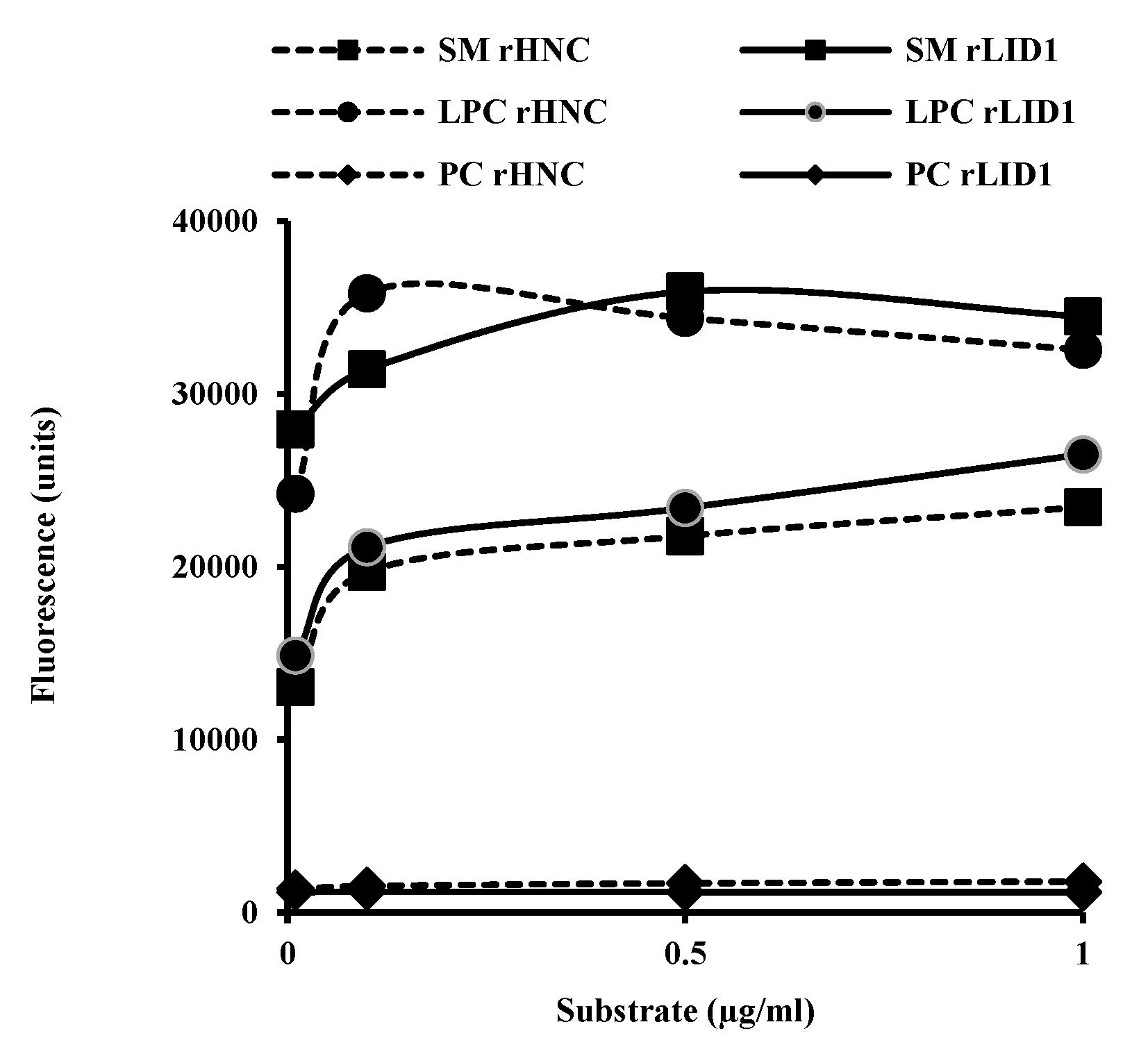
2.3. Phospholipid Substrates Preference of rHNC and rLiD1
2.4. Transphosphatidylation Potential of rHNC and rLiD1
2.5. Similarities and Differences between rLiD1 and rHNC Revealed by Structural Analysis
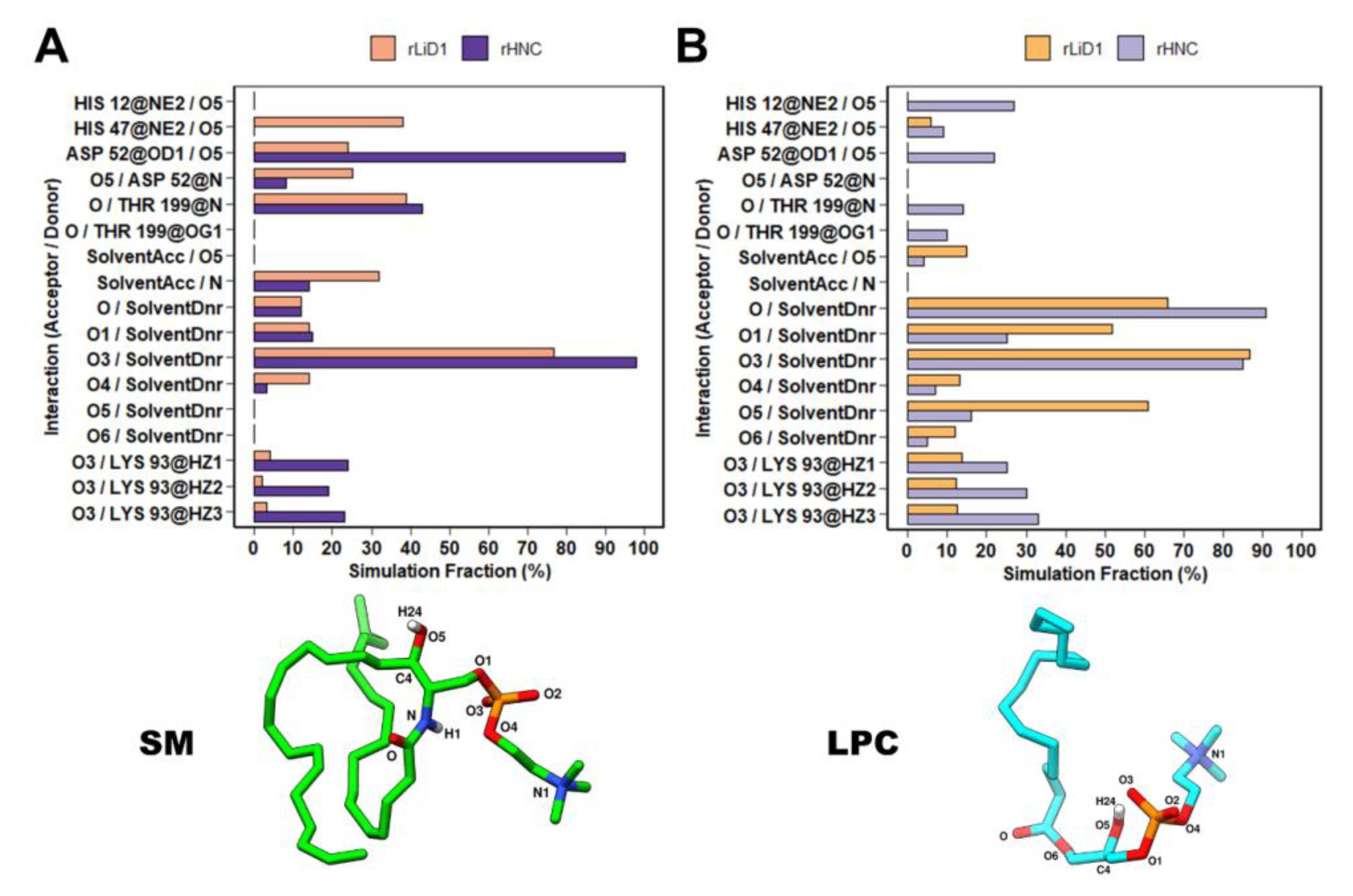
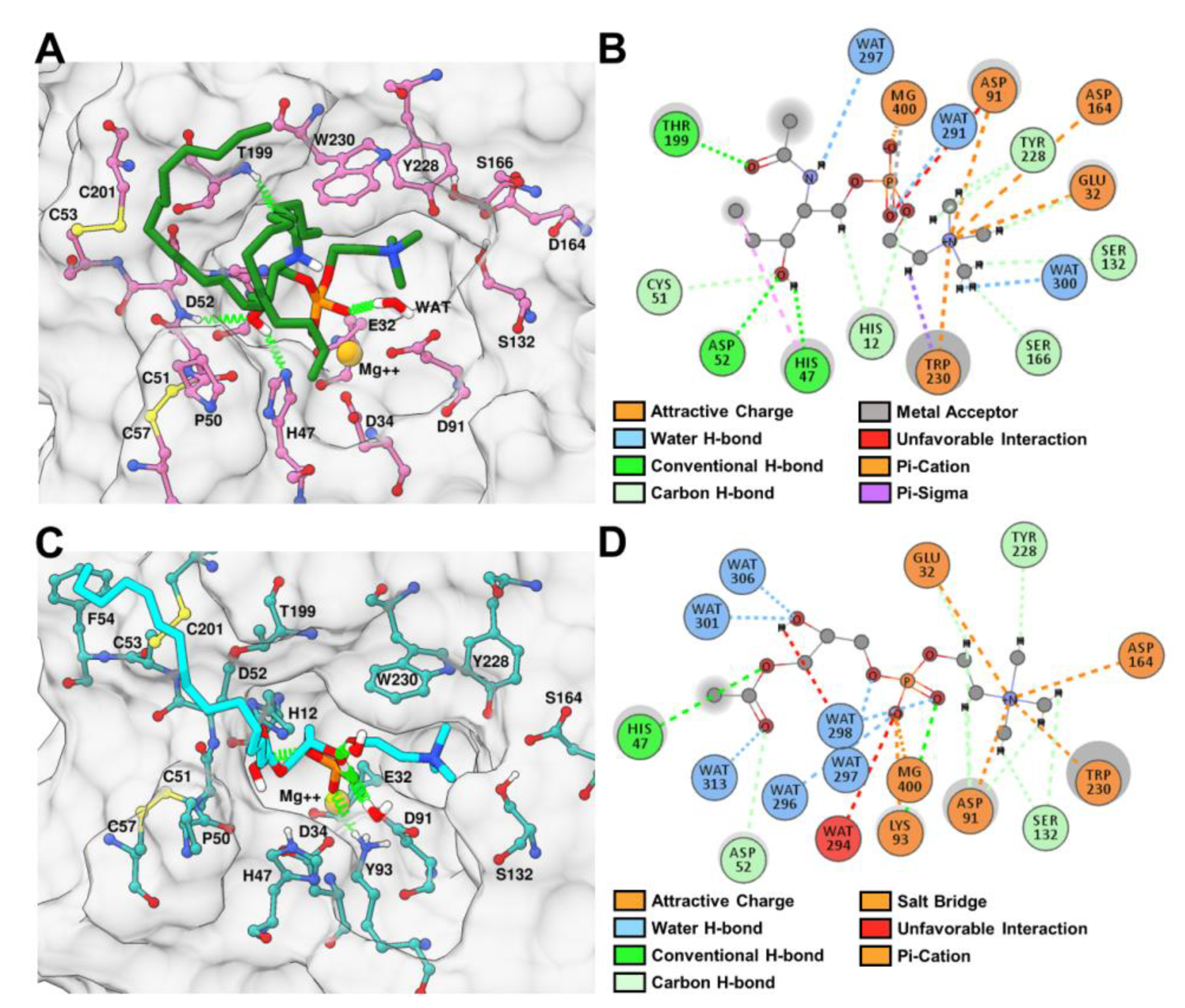
2.6. Binding and Interaction Predictions of SM and LPC with rLiD1 and rHNC
2.7. Dynamic Behavior of rLiD1 and rHNC Structures in Bound and Unbound Simulations
2.8. Dynamic Behavior of Substrates Bound to rLiD1 and rHNC
2.9. rLiD1 and rHNC Intermolecular Interactions with the Substrates
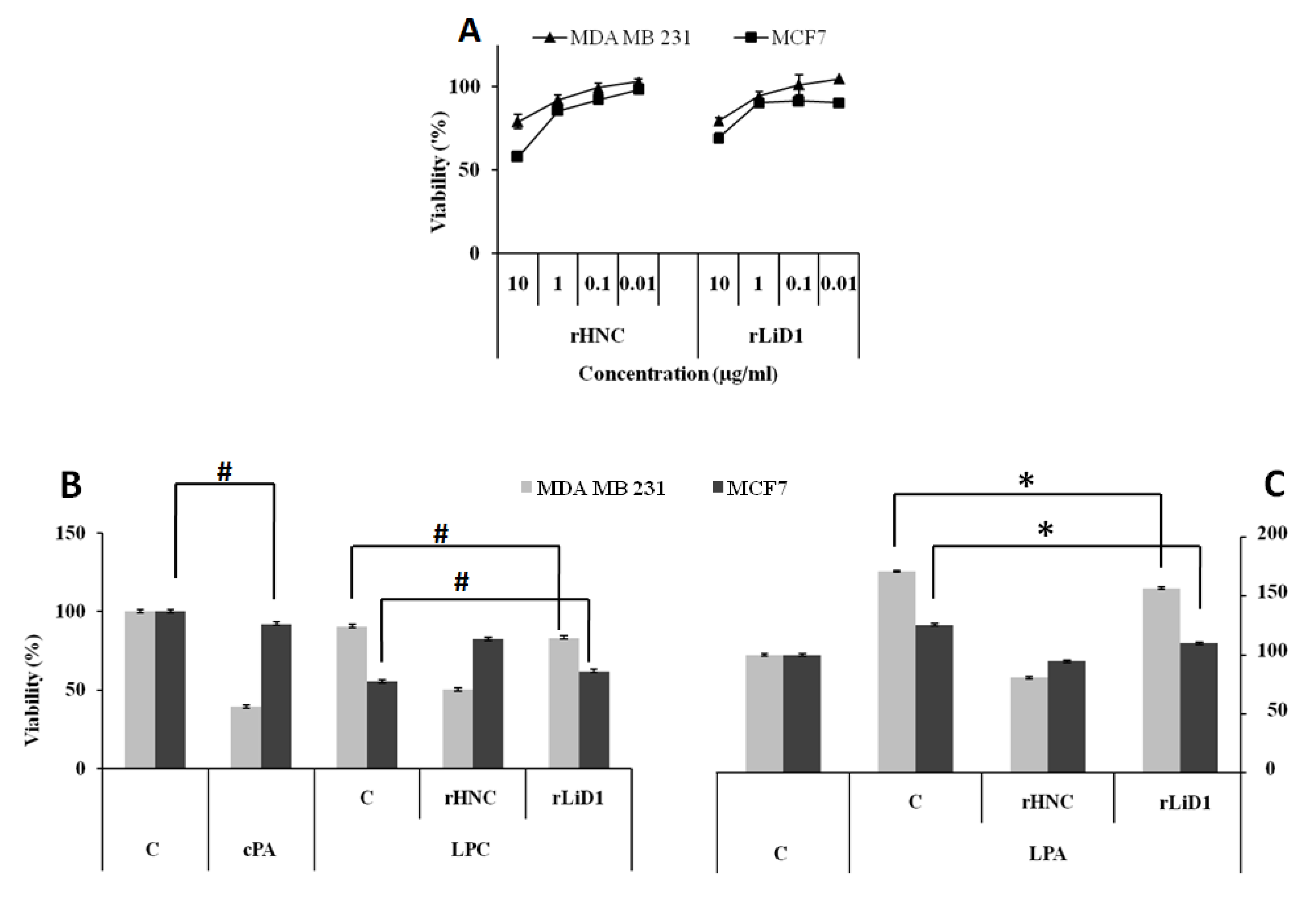
2.10. Cytotoxicity Effect of HNC and rLiD1 on MDA-MB-231 and MCF7 Cell Lines
2.11. Antiproliferative Potential of HNC and rLiD1 on MDA-MB-231 and MCF7 Cell Lines
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Toxins and Venom
5.2. Anti-Venoms and Anti-Toxins
5.3. Immunoassays
5.4. Complement-Dependent Hemolytic Activity
5.5. Cross Inhibition of Hemolysis
5.6. Enzymatic Activities
5.6.1. PLD Activity of rHNC and rLiD1: Choline Release Assay
5.6.2. Thin Layer Chromatography: Detection of Lipids Products from rHNC and rLiD1 Toxins Enzymatic Activity
5.7. Structural Studies
5.7.1. Comparative Modeling
5.7.2. Molecular Docking
5.7.3. Molecular Dynamics
5.8. Cellular Studies
5.8.1. Cell Culture
5.8.2. Viability
5.8.3. Anti-Proliferative Potential Determination of rHNC and rLiD1
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Radmanesh, M. Clinical study of Hemiscorpius lepturus sting in Iran. Am. J. Trop. Med. Hyg. 1990, 93, 327–332. [Google Scholar]
- Ministério da Saúde, Brasil. Banco de dados do Sistema Único de Saúde—DATASUS, Informações de Saúde, Rede Assistencial. 2020. Available online: http://tabnet.datasus.gov.br/cgi/tabcgi.exe?sinannet/cnv/animaisbr.def (accessed on 14 August 2020).
- Jalali, A.; Pipelzadeh, M.H.; Seyedian, R.; Rowan, E.G. A review of epidemiological, clinical and in vitro physiological studies of envenomation by the scorpion Hemiscorpiuslepturus (Hemiscorpiidae) in Iran. Toxicon 2010, 55, 173–179. [Google Scholar] [CrossRef] [PubMed]
- Dias-Lopes, C.; Felicori, L.; Guimarães, G.; Gomes, E.R.; Roman-Campos, D.; Duarte, H.; Damasceno, D.; Martins, M.; Kalapothakis, E.; Almeida, A.P.; et al. Cardiotoxic Effects of Loxosceles Intermedia Spider Venom and the Recombinant Venom Toxin rLiD1. Toxicon 2010, 56, 1426–1435. [Google Scholar] [CrossRef] [PubMed]
- Tambourgi, D.V.; Magnoli, F.C.; van den Berg, C.W.; Morgan, B.P.; de Araujo, P.S.; Alves, E.W.; da Silva, W.D. Sphingomyelinases in the venom of the spider Loxosceles intermedia are responsible for both dermonecrosis and complement-dependent hemolysis. Biochem. Biophys. Res. Commun. 1998, 251, 366–373. [Google Scholar] [CrossRef]
- Borchani, L.; Sassi, A.; Shahbazzadeh, D.; Strub, J.M.; Tounsi, H.; Boubaker, M.S.; Akbari, A.; Van Dorsselaer, A.; El Ayeb, M. Heminecrolysin, the first hemolytic dermonecrotic toxin purified from scorpion venom. Toxicon 2011, 58, 130–139. [Google Scholar] [CrossRef]
- Kurpiewski, G.; Forrester, L.J.; Barrett, J.T.; Campbell, B.J. Platelet aggregation and sphingomyelinase D activity of a purified toxin from the venom of Loxosceles recluse. Biochimica Biophysica Acta 1981, 678, 467–476. [Google Scholar] [CrossRef]
- Van Meeteren, L.A.; Frederiks, F.; Giepmans, B.N.; Fernandes Pedrosa, M.F.; Billington, S.J.; Jost, B.H.; Tambourgi, D.V.; Moolenaar, W.H. Spider and bacterial sphingomyelinases D target cellular LPA receptors by hydrolyzing lysophosphatidylcholine. J. Biol. Chem. 2004, 279, 10833–10836. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.; Lynch, K.R. Brown recluse spider (Loxosce lesreclusa) venom phospholipase D (PLD) generates lysophosphatidic acid (LPA). Biochem. J. 2005, 391, 317–323. [Google Scholar] [CrossRef] [Green Version]
- Lajoie, D.M.; Zobel-Thropp, P.A.; Kumirov, V.K.; Bandarian, V.; Binford, G.J.; Cordes, M.H. Phospholipase D Toxins of Brown Spider Venom Convert Lysophosphatidylcholine and Sphingomyelin to Cyclic Phosphates. PLoS ONE 2013, 29, 8. [Google Scholar] [CrossRef] [Green Version]
- Murakami, M.T.; Fernandes-Pedrosa, M.D.F.; De Andrade, S.A.; Gabdoulkhakov, A.; Betzel, C.; Tambourgi, D.V.; Arni, R.K. Structural insights into the catalytic mechanism of sphingomyelinases D and evolutionary relationship to glycerophosphodiester phosphodiesterases. Biochem. Biophys. Res. Commun. 2006, 342, 323–329. [Google Scholar] [CrossRef]
- De Giuseppe, P.O.; Ullah, A.; Silva, D.T.; Gremski, L.H.; Wille, A.C.; Chaves Moreira, D.; Ribeiro, A.S.; Chaim, O.M.; Murakami, M.T.; Veiga, S.S.; et al. Structure of a novel class II phospholipase D: Catalytic cleft is modified by a disulfide bridge. Biochem. Biophysic. Res. Commun. 2011, 409, 622–627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalapothakis, E.; Chatzaki, M.; Gonçalves-Dornelas, H.; de Castro, C.S.; Silvestre, F.G.; Laborne, F.V.; de Moura, J.F.; Veiga, S.S.; Chávez-Olórtegui, C.; Granier, C.; et al. The Loxtox protein family in Loxosceles intermedia (Mello-Leitão) venom. Toxicon 2007, 50, 938–946. [Google Scholar] [CrossRef] [PubMed]
- Felicori, L.; Araujo, S.C.; de Avila, R.A.; Sanchez, E.F.; Granier, C.; Kalapothakis, E.; Chávez-Olórtegui, C. Functional characterization and epitope analysis of a recombinant dermonecrotic protein from Loxosceles intermedia spider. Toxicon 2006, 48, 509–519. [Google Scholar] [CrossRef]
- Oliveira, C.F.; Vilela, A.; Coura, L.A.; Rodrigues, F.T.; Nagem, R.A.; Chávez-Olortegui, C.; Maioli, T.U.; Felicori, L.F. Protective antibodies against a sphingomyelinase D from Loxosceles intermedia spider venom elicited in mice with different genetic background. Vaccine 2016, 34, 3828–3834. [Google Scholar] [CrossRef] [PubMed]
- Felicori, L.; Fernandes, P.B.; Giusta, M.S.; Duarte, C.G.; Kalapothakis, E.; Nguyen, C.; Molina, F.; Granier, C.; Chávez-Olórtegui, C. An in vivo protective response against toxic effects of the dermonecrotic protein from Loxosceles intermedia spider venom elicited by synthetic epitopes. Vaccine 2009, 27, 4201–4208. [Google Scholar] [CrossRef]
- Mendes, T.M.; Oliveira, D.; Figueiredo, L.F.; Machado-de-Avila, R.A.; Duarte, C.G.; Dias-Lopes, C.; Guimarães, G.; Felicori, L.; Minozzo, J.C.; Chávez-Olortegui, C. Generation and characterization of a recombinant chimeric protein (rCpLi) consisting of B-cell epitopes of a dermonecrotic protein from Loxosceles intermedia spider venom. Vaccine 2013, 31, 2749–2755. [Google Scholar] [CrossRef] [Green Version]
- Souza, N.A.; Dias-Lopes, C.; Matoso, Í.H.G.; de Oliveira, C.F.B.; Chávez-Olortegui, C.D.; Minozzo, J.C.; Felicori, L.F. Immunoprotection elicited in rabbit by a chimeric protein containing B-cell epitopes of Sphingomyelinases D from Loxosceles spp. spiders. Vaccine 2018, 36, 7324–7330. [Google Scholar] [CrossRef]
- Borchani, L.; Sassi, A.; Ben Gharsa, H.; Safra, I.; Shahbazzadeh, D.; Ben Lasfar, Z.; El Ayeb, M. The pathological effects of Heminecrolysin, a dermonecrotic toxin from Hemiscorpius lepturus scorpion venom are mediated through its lysophospholipase D activity. Toxicon 2013, 68, 30–39. [Google Scholar] [CrossRef]
- Borchani, L.; Sassi, A.; Ben Yekhlef, R.; Safra, I.; El Ayeb, M. Heminecrolysin, a potential immunogen for monospecific antivenom production against Hemiscorpius lepturus scorpion. Toxicon 2011, 58, 681–688. [Google Scholar] [CrossRef]
- Torabi, E.; Asgari, S.; Khalaj, V.; Behdani, M.; Kazemi-Lomedasht, F.; Bagheri, K.P.; Shahbazzadeh, D. Corrigendum to “The first report on transcriptome analysis of the venom gland of Iranian scorpion, Hemiscorpius lepturus” [Toxicon 2017, 125, 123–130]. Toxicon 2017, 128, 60. [Google Scholar] [CrossRef]
- Torabi, E.; Behdani, M.; Chafi, M.H.; Moazzami, R.; Sabatier, J.M.; Khalaj, V.; Shahbazzadeh, D.; Bagheri, K.P. Characteristics and Lethality of a Novel Recombinant Dermonecrotic Venom Phospholipase D from Hemiscorpius lepturus. Toxins 2017, 9, 102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lajoie, D.M.; Roberts, S.A.; Zobel-Thropp, P.A.; Delahaye, J.L.; Bandarian, V.; Binford, G.J.; Cordes, M.H. Variable Substrate Preference among Phospholipase D Toxins from Sicariid Spiders. J. Biol. Chem. 2015, 290, 10994–11007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramada, J.S.; Becker-Finco, A.; Minozzo, J.C.; Felicori, L.F.; Machado de Avila, R.A.; Molina, F.; Nguyen, C.; de Moura, J.; Chávez-Olórtegui, C.; Alvarenga, L.M. Synthetic peptides for in vitro evaluation of the neutralizing potency of Loxosceles antivenoms. Toxicon 2013, 73, 47–55. [Google Scholar] [CrossRef] [PubMed]
- De Almeida, D.M.; Gondo-Higashi, H.; Marcelino, J.R.; Ho, P.L.; Junqueira-De-Azevedo, I.D.L.; Berg, C.V.D.; De Andrade, R.M.G.; Tambourgi, D.V.; Fernandes-Pedrosa, M.D.F. A new anti-loxoscelic serum produced against recombinant sphingomyelinase D:Results of preclinical trials. Am. J. Trop. Med. Hyg. 2008, 79, 463–470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murakami, M.T.; Fernandes-Pedrosa, M.F.; Tambourgi, D.V.; Arni, R.K. Structural basis for metal ion coordination and the catalytic mechanism of sphingomyelinases D. J. Biol. Chem. 2005, 280, 13658–13664. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vuitika, L.; Chaves-Moreira, D.; Caruso, I.; Lima, M.A.; Matsubara, F.H.; Murakami, M.T.; Takahashi, H.K.; Toledo, M.S.; Coronado, M.A.; Nader, H.B.; et al. Active Site Mapping of Loxosceles Phospholipases D: Biochemical and Biological Features. Biochim. Biophys. Acta 2016, 1861, 970–979. [Google Scholar] [CrossRef]
- Aoki, J.; Taira, A.; Takanezawa, Y.; Kishi, Y.; Hama, K.; Kishimoto, T.; Mizuno, K.; Saku, K.; Taguchi, R.; Arai, H. Serum Lysophosphatidic Acid Is Produced through Diverse Phospholipase Pathways. J. Biol. Chem. 2002, 277, 48737–48744. [Google Scholar] [CrossRef] [Green Version]
- Ohkawa, R.; Hisano, N.; Nakamura, K.; Okubo, S.; Yokota, H.; Yatomi, Y. Lysophospholipase D activity exists in the urine to catalyze the formation of lysophosphatidic acid. Nephrol. Dial. Transpl. 2006, 21, 3612–3613. [Google Scholar] [CrossRef] [Green Version]
- Moolenaar, W.H. Bioactive Lysophospholipids and Their G Protein-Coupled Receptors. Exp. Cell Res. 1999, 253, 230–238. [Google Scholar] [CrossRef]
- Chang, M.-C.; Lee, J.-J.; Chen, Y.-J.; Lin, S.-I.; Lin, L.-D.; Liou, E.J.-W.; Huang, W.-L.; Chan, C.-P.; Huang, C.-C.; Jeng, J.-H. Lysophosphatidylcholine induces cytotoxicity/apoptosis and IL-8 production of human endothelial cells: Related mechanisms. Oncotarget 2017, 8, 106177–106189. [Google Scholar] [CrossRef] [Green Version]
- Tokumura, A. Physiological and Pathophysiological Roles of Lysophosphatidic Acids Produced by Secretory Lysophospholipase D in Body Fluids. Bioch. Biophys Acta 2002, 1582, 18–25. [Google Scholar] [CrossRef]
- Murakami-Murofushi, K.; Kaji, K.; Kano, K.; Fukuda, M.; Shioda, M.; Murofushi, H. Inhibition of Cell Proliferation by a Unique Lysophosphatidic Acid, PHYLPA, Isolated from PhysarumPolycephalum: Signaling Events of Antiproliferative Action by PHYLPA. Cell Struct. Funct. 1993, 18, 363–370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mukai, M.; Imamura, F.; Ayaki, M.; Shinkai, K.; Iwasaki, T.; Murakami-Murofushi, K.; Murofushi, H.; Kobayashi, S.; Yamamoto, T.; Nakamura, H.; et al. Inhibition of tumor invasion and metastasis by a novel lysophosphatidic acid (cyclic LPA). Int. J. Cancer 1999, 81, 918–922. [Google Scholar] [CrossRef]
- Murakami-Murofushi, K.; Uchiyama, A.; Fujiwara, Y.; Kobayashi, T.; Kobayashi, S.; Mukai, M.; Murofushi, H.; Tigyi, G. Biological Functions of a Novel Lipid Mediator, Cyclic Phosphatidic Acid. Biochim. Biophys. Acta 2002, 1582, 1–7. [Google Scholar] [CrossRef]
- Simstein, R.; Burow, M.; Parker, A.; Weldon, C.; Beckman, B. Apoptosis, chemoresistance, and breast cancer: Insights from the MCF-7 cell model system. Exp. Biol. Med. 2003, 228, 995–1003. [Google Scholar] [CrossRef]
- Paixão-Cavalcante, D.; van den Berg, C.W.; de Freitas Fernandes-Pedrosa, M.; Gonçalves de Andrade, R.M.; Tambourgi, D.V. Role of matrix metalloproteinases in HaCaT keratinocytes apoptosis induced by Loxosceles venom sphingomyelinase D. J. Investig. Dermatol. 2006, 126, 61–68. [Google Scholar] [CrossRef] [Green Version]
- Lopes, P.H.; Berg, C.W.; Tambourgi, D.V. Sphingomyelinases D from Loxosceles Spider Venoms and Cell Membranes: Action on Lipid Rafts and Activation of Endogenous Metalloproteinases. Front. Pharmacol. 2020, 11, 636. [Google Scholar] [CrossRef]
- Tokumura, A.; Majima, E.; Kariya, Y.; Tominaga, K.; Kogure, K.; Yasuda, K.; Fukuzawa, K. Identification of human plasma lysophospholipase D, a lysophosphatidic acid-producing enzyme, as autotaxin, a multifunctional phosphodiesterase. J. Biol. Chem. 2002, 277, 39436–39442. [Google Scholar] [CrossRef] [Green Version]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [Green Version]
- Di Tommaso, P.; Moretti, S.; Xenarios, I.; Orobitg, M.; Montanyola, A.; Chang, J.M.; Taly, J.F.; Notredame, C. T-Coffee: A web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res. 2011, 39, 13–17. [Google Scholar] [CrossRef]
- Webb, B.; Sali, A. Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinform. 2014, 47, 5–6. [Google Scholar] [CrossRef] [Green Version]
- Bhattacharya, D.; Nowotny, J.; Cao, R.; Cheng, J. 3Drefine: An interactive web server for efficient protein structure refinement. Nucleic Acids Res. 2016, 44, W406–W409. [Google Scholar] [CrossRef]
- Benkert, P.; Künzli, M.; Schwede, T. QMEAN server for protein model quality estimation. Nucleic Acids Res. 2009. [Google Scholar] [CrossRef] [Green Version]
- Colovos, C.; Yeates, T.O. Verification of protein structures: Patterns of nonbonded atomic interactions. Protein Sci. 1993, 2, 1511–1519. [Google Scholar] [CrossRef] [Green Version]
- Eisenberg, D.; Lüthy, R.; Bowie, J.U. VERIFY3D: Assessment of protein models with three-dimensional profiles. Methods Enzymol. 1997, 277, 396–404. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B. PubChem 2019 update: Improved access to chemical data. Nucleic Acids Res. 2019, 47, D1102–D1109. [Google Scholar] [CrossRef] [Green Version]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [Green Version]
- Biovia, D.S. Discovery Studio Visualizer. 2019. Available online: https://discover.3ds.com/discovery-studio-visualizer (accessed on 14 August 2020).
- Case, D.A.; Betz, R.M.; Cerutti, D.S.; Cheatham Iii, T.E.; Darden, T.A.; Duke, R.E.; Giese, T.J.; Gohlke, H.; Goetz, A.W.; Homeyer, N.; et al. Amber 2016; University of California: San Francisco, CA, USA, 2016; p. 810. [Google Scholar]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef]
- Jakalian, A.; Jack, D.B.; Bayly, C.I. Fast, efficient generation of high-quality atomic charges. AM1-BCC model: II. Parameterization and validation. J. Comput. Chem. 2002, 23, 1623–1641. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, W.; Kollman, P.A.; Case, D.A. Automatic atom type and bond type perception in molecular mechanical calculations. J. Mol. Graph. Model. 2006, 25, 247–260. [Google Scholar] [CrossRef] [PubMed]
- Dolinsky, T.J.; Nielsen, J.E.; McCammon, J.A.; Baker, N.A. PDB2PQR: An automated pipeline for the setup of Poisson–Boltzmann electrostatics calculations. Nucleic Acids Res. 2004, 32, W665–W667. [Google Scholar] [CrossRef]
- Case, D.A.; Ben-Shalom, I.Y.; Brozell, S.R.; Cerutti, D.S.; Cheatham III, T.E.; Cruzeiro, V.W.D.; Darden, T.A.; Duke, R.E.; Ghoreishi, D.; Gilson, M.K.; et al. Amber 2018; University of California: San Francisco, CA, USA, 2018. [Google Scholar]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Development Core Team, R. R: A Language and Environment for Statistical Computing, reference index version 2.15.1 R foundation for statistical Computing, Vienne. 2013. Available online: https://www.r-project.org/ (accessed on 7 November 2019).

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ben Yekhlef, R.; Felicori, L.; Santos, L.H.; F. B. Oliveira, C.; Fadhloun, R.; Torabi, E.; Shahbazzadeh, D.; Pooshang Bagheri, K.; Salgado Ferreira, R.; Borchani, L. Antigenic and Substrate Preference Differences between Scorpion and Spider Dermonecrotic Toxins, a Comparative Investigation. Toxins 2020, 12, 631. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins12100631
Ben Yekhlef R, Felicori L, Santos LH, F. B. Oliveira C, Fadhloun R, Torabi E, Shahbazzadeh D, Pooshang Bagheri K, Salgado Ferreira R, Borchani L. Antigenic and Substrate Preference Differences between Scorpion and Spider Dermonecrotic Toxins, a Comparative Investigation. Toxins. 2020; 12(10):631. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins12100631
Chicago/Turabian StyleBen Yekhlef, Ramla, Liza Felicori, Lucianna Helene Santos, Camila F. B. Oliveira, Raoudha Fadhloun, Elham Torabi, Delavar Shahbazzadeh, Kamran Pooshang Bagheri, Rafaela Salgado Ferreira, and Lamia Borchani. 2020. "Antigenic and Substrate Preference Differences between Scorpion and Spider Dermonecrotic Toxins, a Comparative Investigation" Toxins 12, no. 10: 631. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins12100631