A Novel Microbial Zearalenone Transformation through Phosphorylation
Abstract
:1. Introduction
2. Results
2.1. Isolation of ZEA Transforming Bacteria
2.2. Taxonomic Identity of the Active Isolate S62-W
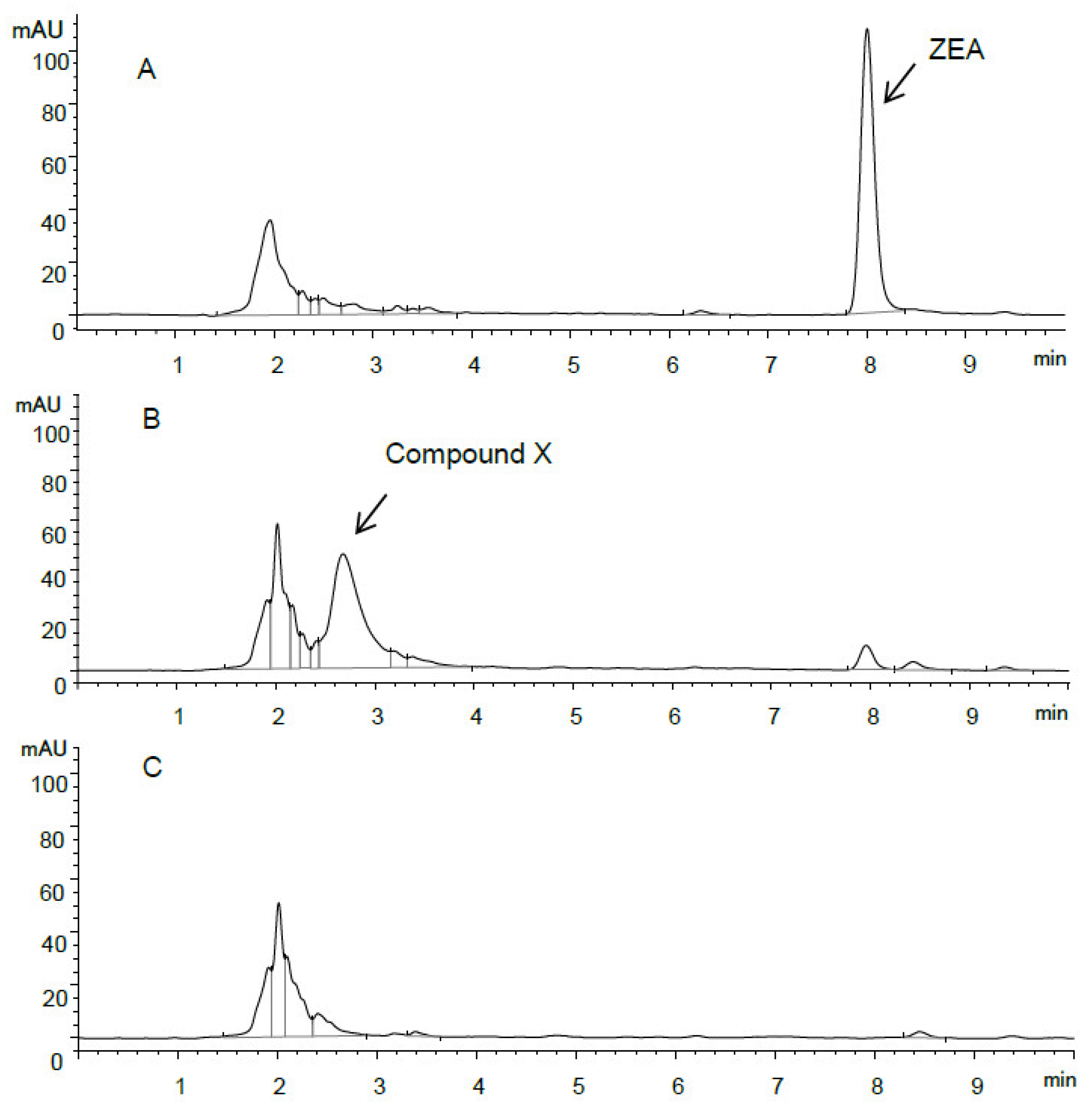
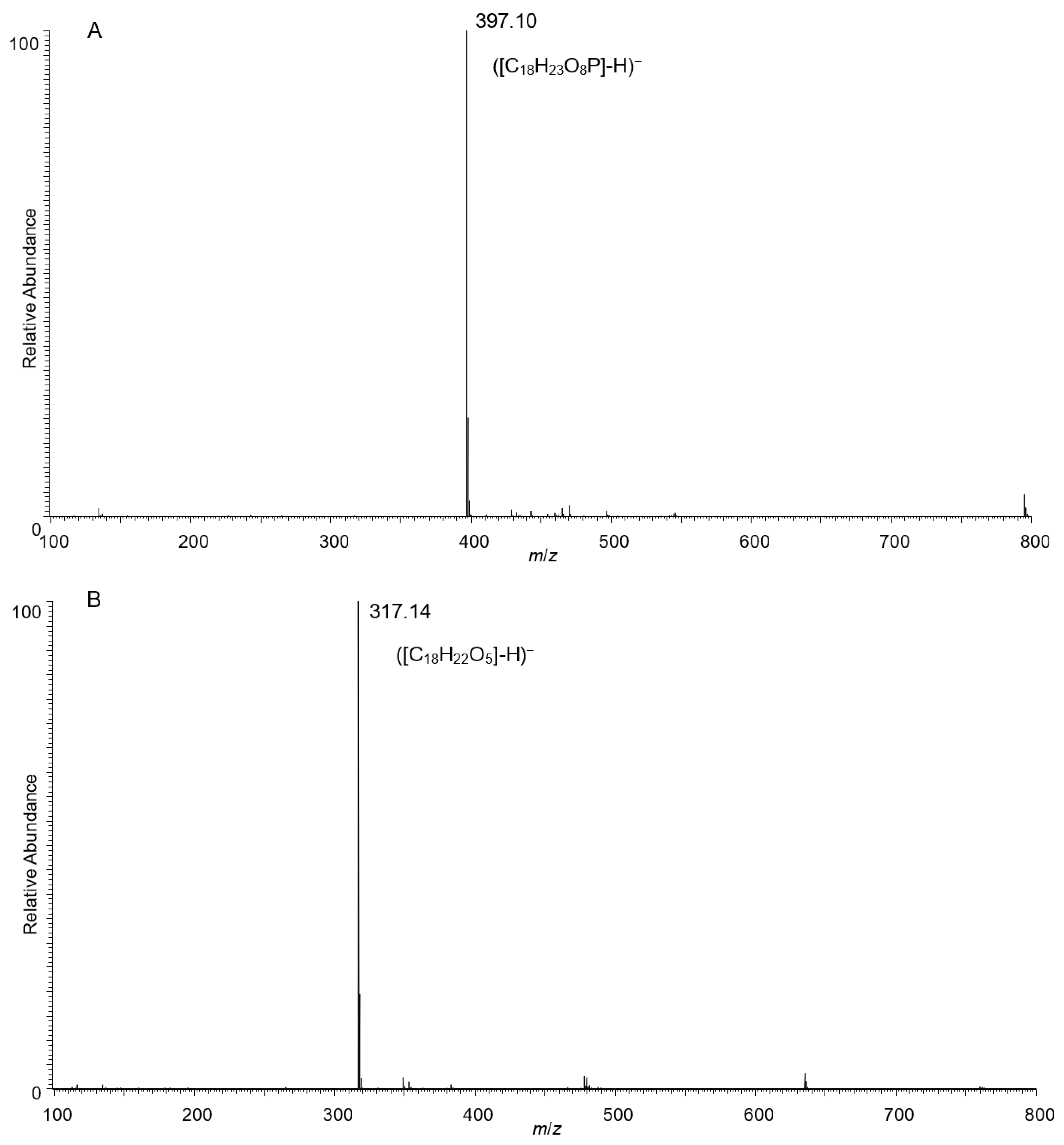
2.3. Identification of ZEA Transformation Product
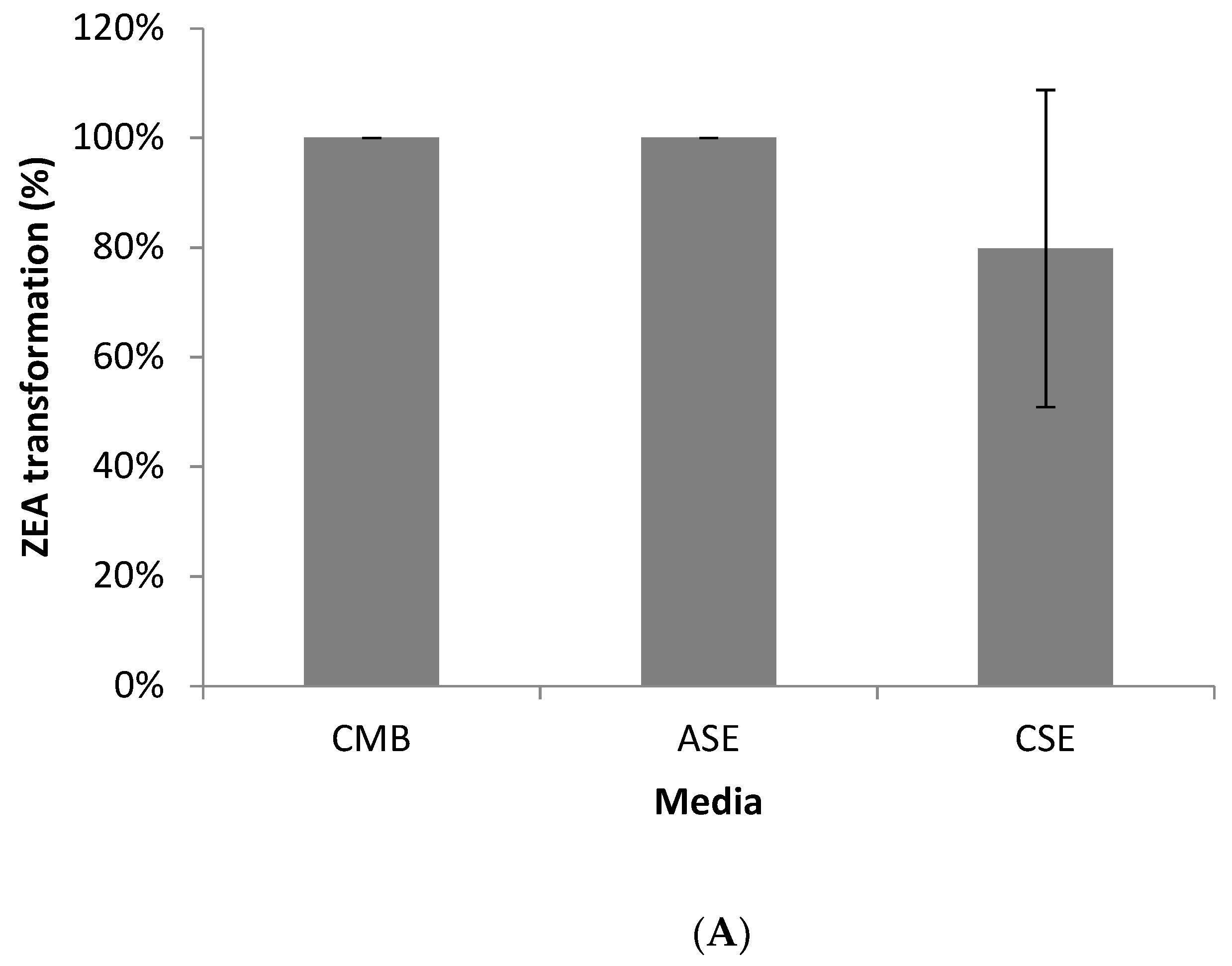
2.4. ZEA Transformation by Bacillus sp. S62-W in Media
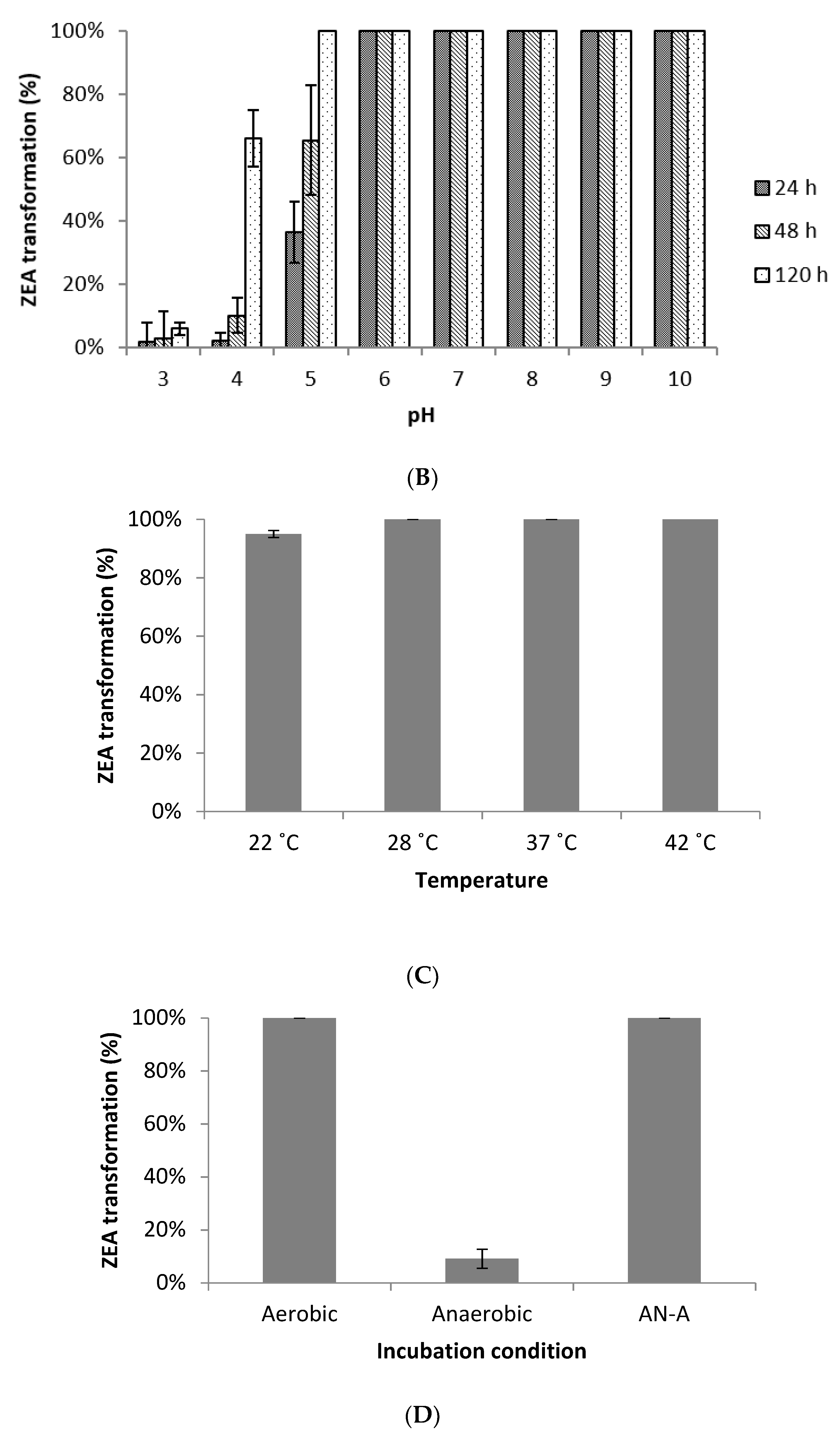
2.5. Effects of Environmental Factors on ZEA Transformation
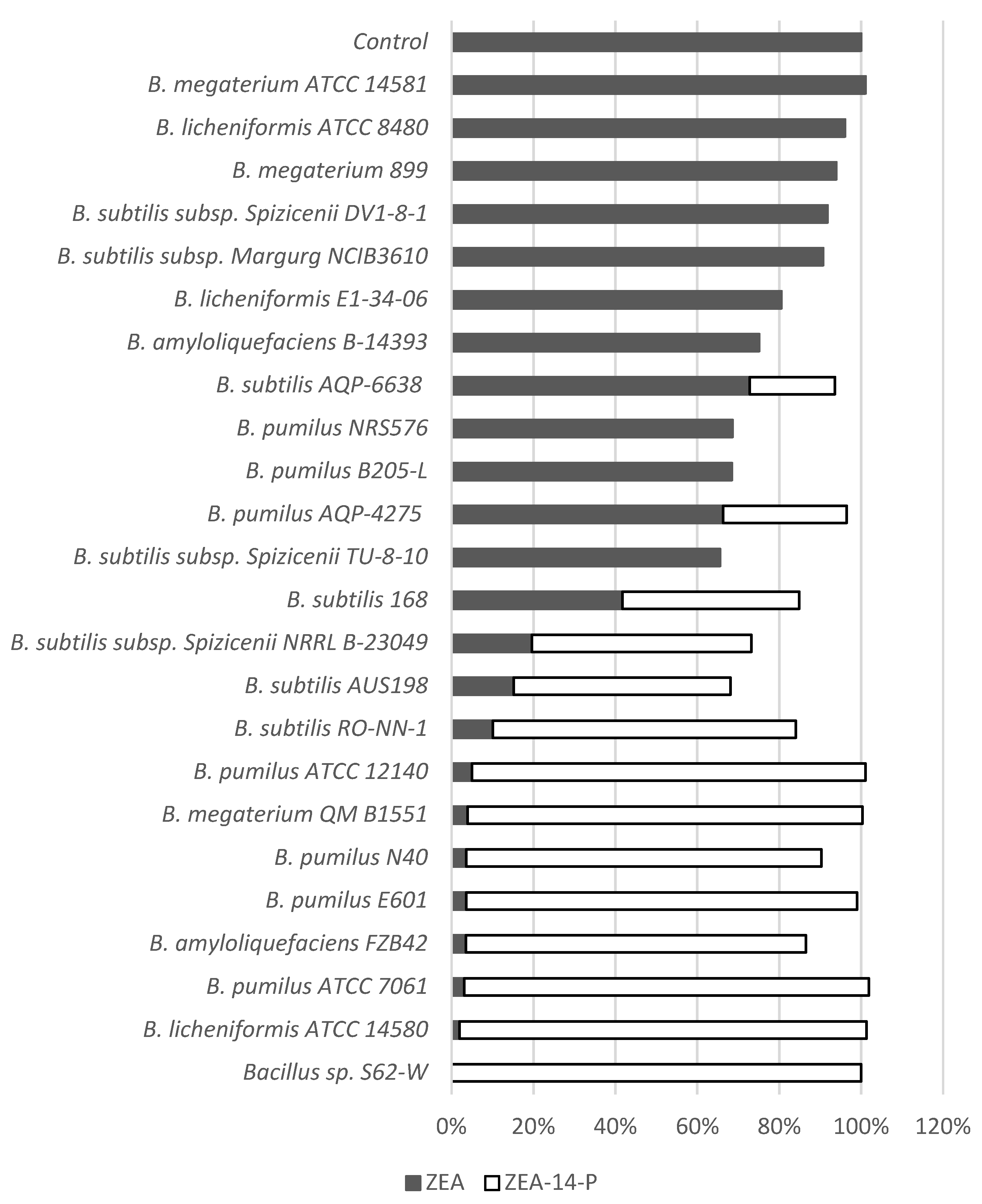
2.6. Determination of ZEA Phosphorylation in Various Bacillus Strains
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Chemicals and Culture Media
5.2. Screening of Corn Silage Samples
5.3. Single Colony Isolation and Purification
5.4. Taxonomic Identification of the Active Isolate
5.5. ZEA Transformation Activities of the Active Isolate
5.6. Analysis of ZEA by HPLC
5.7. Extraction and Purification of ZEA Transformation Product
5.8. Identification of ZEA Transformation Product by LC-MS/MS and NMR
5.9. Identification of ZEA Transformation Product by Comparing Standard ZEA-14-Phosphate and ZEA-16-Phosphate
5.10. Determination of ZEA Phosphorylation in Various Bacillus Strains
5.11. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zinedine, A.; Soriano, J.M.; Moltó, J.C.; Mañes, J. Review on the toxicity, occurrence, metabolism, detoxification, regulations and intake of zearalenone: An oestrogenic mycotoxin. Food Chem. Toxicol. 2007, 45, 1–18. [Google Scholar] [CrossRef]
- Tanaka, T.; Hasegawa, A.; Yamamoto, S.; Lee, U.S.; Sugiura, Y.; Ueno, Y. Worldwide contamination of cereals by the fusarium mycotoxins nivalenol, deoxynivalenol, and zearalenone. 1. Survey of 19 countries. J. Agric. Food Chem. 1988, 36, 979–983. [Google Scholar] [CrossRef]
- Tangni, E.K.; Pussemier, L.; Van Hove, F. Mycotoxin contaminating maize and grass silages for dairy cattle feeding: Current state and challenges. J. Anim. Sci. Adv. 2013, 3, 492–511. [Google Scholar]
- Ogunade, I.M.; Martinez-Tuppia, C.; Queiroz, O.C.M.; Jiang, Y.; Drouin, P.; Wu, F.; Vyas, D.; Adesogan, A.T. Silage review: Mycotoxins in silage: Occurrence, effects, prevention, and mitigation. J. Dairy Sci. 2018, 101, 4034–4059. [Google Scholar] [CrossRef] [PubMed]
- EFSA. Scientific opinion on the risks for public health related to the presence of zearalenone in food. EFSA J. 2011, 9, 2197. [Google Scholar] [CrossRef]
- Pereyra, M.L.G.; Sulyok, M.; Baralla1, V.; Dalcero1, A.M.; Krska, R.; Chulze, S.; Cavaglieri, L.R. Evaluation of zearalenone, α-zearalenol, β-zearalenol, zearalenone 4-sulfate and β-zearalenol 4-glucoside levels during the ensiling process. World Mycotoxin J. 2014, 7, 291–295. [Google Scholar] [CrossRef]
- He, J.; Zhou, T. Patented techniques for detoxification of mycotoxins in feeds and food matrices. Recent Pat. Food Nutr. Agric. 2010, 2, 96–104. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Hassan, Y.I.; Watts, C.; Zhou, T. Innovative technologies for the mitigation of mycotoxins in animal feed and ingredients—A review of recent patents. Anim. Feed Sci. Technol. 2016, 216, 19–29. [Google Scholar] [CrossRef]
- Yi, P.; Pai, C.; Liu, J. Isolation and characterization of a bacillus licheniformis strain capable of degrading zearalenone. World J. Microbiol. Biotechnol. 2011, 27, 1035–1043. [Google Scholar] [CrossRef]
- Cho, K.J.; Kang, J.S.; Cho, W.T.; Lee, C.H.; Ha, J.K.; Song, K.B. In vitro degradation of zearalenone by bacillus subtilis. Biotechnol. Lett. 2010, 32, 1921–1924. [Google Scholar] [CrossRef]
- Lei, Y.; Zhao, L.; Ma, Q.; Zhang, J.; Zhou, T.; Gao, C.; Ji, C. Degradation of zearalenone in swine feed and feed ingredients by bacillus subtilis ansb01g. World Mycotoxin J. 2014, 7, 143–151. [Google Scholar] [CrossRef]
- Tinyiro, S.E.; Yao, W.; Sun, X.; Wokadala, C.; Wang, S. Scavenging of zearalenone by bacillus strains-in vitro. Res. J. Microbiol. 2011, 6, 304–309. [Google Scholar] [CrossRef] [Green Version]
- Tinyiro, S.E.; Wokadala, C.; Xu, D.; Yao, W. Adsorption and degradation of zearalenone by bacillus strains. Folia Microbiol. 2011, 56, 321–327. [Google Scholar] [CrossRef]
- Tan, H.; Hu, Y.; He, J.; Wu, L.; Liao, F.; Luo, B.; He, Y.; Zuo, Z.; Ren, Z.; Zhong, Z.; et al. Zearalenone degradation by two pseudomonas strains from soil. Mycotoxin Res. 2014, 30, 191–196. [Google Scholar] [CrossRef] [PubMed]
- Tan, H.; Zhang, Z.; Hu, Y.; Wu, L.; Liao, F.; He, J.; Luo, B.; He, Y.; Zuo, Z.; Ren, Z.; et al. Isolation and characterization of pseudomonas otitidis th-n1 capable of degrading zearalenone. Food Control 2015, 47, 285–290. [Google Scholar] [CrossRef]
- Yu, Y.; Qiu, L.; Wu, H.; Tang, Y.; Yu, Y.; Li, X.; Liu, D. Degradation of zearalenone by the extracellular extracts of acinetobacter sp. Sm04 liquid cultures. Biodegradation 2011, 22, 613–622. [Google Scholar] [CrossRef] [PubMed]
- Kriszt, R.; Krifaton, C.; Szoboszlay, S.; Cserháti, M.; Kriszt, B.; Kukolya, J.; Czéh, A.; Fehér-Tóth, S.; Török, L.; Szőke, Z.; et al. A new zearalenone biodegradation strategy using non-pathogenic rhodococcus pyridinivorans k408 strain. PLoS ONE 2012, 7, e43608. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Dong, M.; Yang, Q.; Apaliya, M.T.; Li, J.; Zhang, X. Biodegradation of zearalenone by saccharomyces cerevisiae: Possible involvement of zen responsive proteins of the yeast. J. Proteom. 2016, 143, 416–423. [Google Scholar] [CrossRef]
- Brodehl, A.; Möller, A.; Kunte, H.J.; Koch, M.; Maul, R. Biotransformation of the mycotoxin zearalenone by fungi of the genera rhizopus and aspergillus. FEMS Microbiol. Lett. 2014, 359, 124–130. [Google Scholar] [CrossRef] [Green Version]
- Jard, G.; Liboz, T.; Mathieu, F.; Guyonvarc’h, A.; André, F.; Delaforge, M.; Lebrihi, A. Transformation of zearalenone to zearalenone-sulfate by aspergillus spp. World Mycotoxin J. 2010, 3, 183–191. [Google Scholar] [CrossRef]
- Sun, X.; He, X.; Xue, K.; Li, Y.; Xu, D.; Qian, H. Biological detoxification of zearalenone by aspergillus niger strain fs10. Food Chem. Toxicol. 2014, 72, 76–82. [Google Scholar] [CrossRef]
- El-Sharkawy, S.H.; Selim, M.I.; Afifi, M.S.; Halaweish, F.T. Microbial transformation of zearalenone to a zearalenone sulfate. Appl. Environ. Microbiol. 1991, 57, 549–552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vekiru, E.; Hametner, C.; Mitterbauer, R.; Rechthaler, J.; Adam, G.; Schatzmayr, G.; Krska, R.; Schuhmacher, R. Cleavage of zearalenone by trichosporon mycotoxinivorans to a novel nonestrogenic metabolite. Appl. Environ. Microbiol. 2010, 76, 2353–2359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kakeya, H.; Takahashi-Ando, N.; Kimura, M.; Onose, R.; Yamaguchi, I.; Osada, H. Biotransformation of the mycotoxin, zearalenone, to a non-estrogenic compound by a fungal strain of clonostachys sp. Biosci. Biotechnol. Biochem. 2002, 66, 2723–2726. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El-Sharkawy, S.; Abul-Hajj, Y. Microbial transformation of zearalenone, i. Formation of zearalenone-4-o-β-glucoside. J. Nat. Prod. 1987, 50, 520–521. [Google Scholar] [CrossRef]
- Branquinho, R.; Meirinhos-Soares, L.; Carriço, J.A.; Pintado, M.; Peixe, L.V. Phylogenetic and clonality analysis of bacillus pumilus isolates uncovered a highly heterogeneous population of different closely related species and clones. FEMS Microbiol. Ecol. 2014, 90, 689–698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Y.; Hassan, Y.I.; Lepp, D.; Shao, S.; Zhou, T. Strategies and methodologies for developing microbial detoxification systems to mitigate mycotoxins. Toxins 2017, 9, 130. [Google Scholar] [CrossRef] [Green Version]
- Mitchell, S.C. Xenobiotic conjugation with phosphate—A metabolic rarity. Xenobiotica 2016, 46, 743–756. [Google Scholar] [CrossRef]
- Day, E.K.; Sosale, N.G.; Lazzara, M.J. Cell Signaling regulation by protein phosphorylation: A multivariate, heterogeneous, and context-dependent process. Curr. Opin. Biotechnol. 2016, 40, 185–192. [Google Scholar] [CrossRef] [Green Version]
- Hsu, C.; Chang, C.; Wang, S.; Fang, T.; Lee, M.; Su, N. Soy isoflavone-phosphate conjugates derived by cultivating bacillus subtilis var. Natto bcrc 80517 with isoflavone. Food Res. Int. 2013, 53, 487–495. [Google Scholar] [CrossRef]
- Abou-Donia, M.B.; Elmasry, E.M.; Abu-Qare, A.W. Metabolism and toxicokinetics of xenobiotics. In Handbook of toxicology, 2nd ed.; Derelanko, M.J., Hollinger, M.A., Eds.; CRC Press: Boca Raton, FL, USA, 2001. [Google Scholar]
- EFSA. Update of the list of qps-recommended biological agents intentionally added to food or feed as notified to efsa 4: Suitability of taxonomic units notified to efsa until March 2016. EFSA J. 2016, 14, 4522. [Google Scholar]
- Wang, W.; Sun, M. Phylogenetic relationships between Bacillus species and related genera inferred from 16s rDNA sequences. Braz. J. Microbiol. 2009, 40, 505–521. [Google Scholar] [CrossRef] [Green Version]
- Te Giffel, M.C.; Wagendorp, A.; Herrewegh, A.; Driehuis, F. Bacterial spores in silage and raw milk. Antonie Leeuwenhoek. 2002, 81, 625–630. [Google Scholar] [CrossRef]
- Ávila, C.L.S.; Carvalho, B.F. Silage fermentation—Updates focusing on the performance of micro-organisms. J. Appl. Microbiol. 2020, 128, 966–984. [Google Scholar] [CrossRef] [Green Version]
- Drouin, P.; Tremblay, J.; Renaud, J.; Apper, E. Microbiota succession during aerobic stability of maize silage inoculated with Lentilactobacillus buchneri NCIMB 40788 and Lentilactobacillus hilgardii CNCM-I-4785. MicrobiologyOpen 2021, 10, e1153. [Google Scholar] [CrossRef] [PubMed]
- Queiroz, O.C.M.; Ogunade, I.M.; Weinberg, Z.; Adesogan, A.T. Silage review: Foodborne pathogens in silage and their mitigation by silage additives. J. Dairy Sci. 2017, 101, 4132–4142. [Google Scholar] [CrossRef] [PubMed]
- Lara Erika, C.; Basso Fernanda, C.; de Assis Flávia, B.; Souza Fernando, A.; Berchielli Telma, T.; Reis Ricardo, A. Changes in the nutritive value and aerobic stability of corn silages inoculated with Bacillus subtilis alone or combined with Lactobacillus Plantarum. Anim. Prod. Sci. 2015, 56, 1867–1874. [Google Scholar] [CrossRef] [Green Version]
- Lee, A.; Cheng, K.C.; Liu, J.R. Isolation and characterization of a Bacillus amyloliquefaciens strain with zearalenone removal ability and its probiotic potential. PLoS ONE 2017, 12, e0182220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Danilova, I.; Sharipova, M. The practical potential of Bacilli and their enzymes for industrial production. Front. Microbiol. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Elshaghabee, F.M.F.; Rokana, N.; Gulhane, R.D.; Sharma, C.; Panwar, H. Bacillus as potential probiotics: Status, concerns, and future perspectives. Front. Microbiol. 2017, 8, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El-Sharkawy, S.H.; Abul-Hajj, Y.J. Microbial transformation of zearalenone. 2. Reduction, hydroxylation, and methylation products. J. Org. Chem. 1988, 53, 515–519. [Google Scholar] [CrossRef]
- Rychlik, M.; Humpf, H.; Marko, D.; Dänicke, S.; Mally, A.; Berthiller, F.; Klaffke, H.; Lorenz, N. Proposal of a comprehensive definition of modified and other forms of mycotoxins including “masked” mycotoxins. Mycotoxin Res. 2014, 30, 197–205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berthiller, F.; Crews, C.; Dall’Asta, C.; Saeger, S.D.; Haesaert, G.; Karlovsky, P.; Oswald, I.P.; Seefelder, W.; Speijers, G.; Stroka, J. Masked mycotoxins: A review. Mol. Nutr. Food Res. 2013, 57, 165–186. [Google Scholar] [CrossRef]
- Gratz, S.W.; Dinesh, R.; Yoshinari, T.; Holtrop, G.; Richardson, A.J.; Duncan, G.; MacDonald, S.; Lloyd, A.; Tarbin, J. Masked trichothecene and zearalenone mycotoxins withstand digestion and absorption in the upper gi tract but are efficiently hydrolyzed by human gut microbiota in vitro. Mol. Nutr. Food Res. 2016. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. Spades: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [Green Version]
- Richter, M.; Rossello-Mora, R.; Oliver Glockner, F.; Peplies, J. Jspeciesws: A web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 2016, 32, 929–931. [Google Scholar] [CrossRef]
- Bertels, F.; Silander, O.K.; Pachkov, M.; Rainey, P.B.; van Nimwegen, E. Automated reconstruction of whole-genome phylogenies from short-sequence reads. Mol. Biol. Evol. 2014, 31, 1077–1088. [Google Scholar] [CrossRef]
- Stamatakis, A. Raxml version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive tree of life (itol) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef] [PubMed]






| Microorganism | Accession No. | Identity (%) |
|---|---|---|
| Bacillus aerius 24K | NR118439.1 | 99 |
| Bacillus stratosphericus 41KF2a | NR042336.1 | 99 |
| Bacillus altitudinis 41KF2b | NR042337.1 | 99 |
| Bacillus pumilus NBRC 12092 | NR112637.1 | 99 |
| Bacillus pumilus SAFR-032 | NR074977.1 | 99 |
| Bacillus pumilus ATCC 7061 | NR043242.1 | 99 |
| Bacillus pumilus SBMP2 | NR118381.1 | 99 |
| Bacillus safensis NBRC 100820 | NR113945.1 | 99 |
| Bacillus safensis FO-366 | NR041794.1 | 99 |
| Microorganism | Z-Scores | ANIb (%) |
|---|---|---|
| Bacillus stratosphericus LK31 | 0.99986 | 98.05 |
| Bacillus stratosphericus (GCA_001043535) LK5 | 0.99984 | 98.07 |
| Bacillus stratosphericus LK23 | 0.99982 | 98.06 |
| Bacillus sp. LK10 | 0.99982 | 98.06 |
| Bacillus stratosphericus (GCA_001038775) LK33 | 0.99981 | 98.06 |
| Bacillus sp. L_1B0_12 | 0.99979 | 98.02 |
| Bacillus stratosphericus (GCA_001038845) LK18 | 0.99979 | 98.05 |
| Bacillus sp. TH007 | 0.99979 | 98.79 |
| Bacillus pumilus (GCA_000828455) B4133 | 0.99975 | 98.30 |
| Bacillus stratosphericus (GCA_001265125) M53 | 0.99975 | 98.09 |
| Bacillus cellulasensis NIO-1130 | 0.99974 | 97.91 |
| Bacillus altitudinis 41KF2b | 0.99973 | 98.01 |
| Bacillus sp. FJAT-21955 | 0.99971 | 98.51 |
| Bacillus pumilus (GCA_000972685) W3 | 0.99965 | 98.10 |
| Atom | Chemical Shift (ppm) | COSY | HSQC | HMBC | ROESY |
|---|---|---|---|---|---|
| 1C | 171.27 | 3, 13, 15 | |||
| 3C | 74.47 | 3 | 5b, 4b, 5a, 19 | ||
| 4C | 35.22 | 4a, 4b | 19, 6a, 6b, 5b, 5a, 3 | ||
| 5C | 22.31 | 5a, 5b | 3, 19, 6a, 4b, 4a, 6b | ||
| 6C | 43.85 | 6a, 6b | 4b, 5b, 5a | ||
| 7C | 216.18 | 6a, 6b, 8b, 8a, 9b, 5b, 9a, 5a | |||
| 8C | 37.68 | 8b, 8a | 6a, 9b, 9a, 6b, 10a, 10b | ||
| 9C | 21.60 | 9b, 9a | 11, 8b, 8a, 10a, 10b, 12 | ||
| 10C | 31.65 | 10b, 10a | 11, 8a, 8b, 9b, 9a, 12 | ||
| 11C | 134.56 | 11 | 12, 9b, 9a, 10b, 10a | ||
| 12C | 131.54 | 12 | 11, 13, 10b, 10a | ||
| 13C | 112.04 | 13 | 12, 15 | ||
| 14C | 157.33 | 13, 15 | |||
| 15C | 107.35 | 15 | 13 | ||
| 16C | 161.26 | 15 | |||
| 17C | 110.12 | 15, 13, 12, 19 | |||
| 18C | 142.00 | 12, 11 | |||
| 19C | 20.41 | 19(H3) | 4b, 3 | ||
| 3H | 5.01 | 19, 4b | 3 | 5, 1, 4, 19 | 5b, 4a, 4b, 5a, 6a |
| 4Ha | 1.61 | 4b | 4 | 5 | 3, 6a |
| 4Hb | 1.51 | 3, 4a, 5a | 4 | 5, 6, 19, 3 | 12, 3, 6a, 6b |
| 5Ha | 1.62 | 5b, 4b, 6b, 6a | 5 | 7, 4, 6, 3 | 3, 8a, 6a |
| 5Hb | 1.70 | 6a, 6b, 5a | 5 | 7, 4, 6, 3 | 12, 3, 19, 8a, 8b, 6a |
| 6Ha | 2.45 | 6b, 5b, 5a | 6 | 7, 8, 4, 5 | 3, 4b, 5b, 5a, 4a |
| 6Hb | 2.17 | 11, 6a, 5b, 5a | 6 | 7, 4, 8, 5 | 8b, 4b |
| 8Ha | 2.38 | 8b, 9b, 9a | 8 | 7, 9, 10 | 11, 9b, 5b, 9a, 5a |
| 8Hb | 2.58 | 8a, 9b, 9a | 8 | 7, 9, 10 | 11, 5b, 6b, 9a |
| 9Ha | 1.63 | 9b, 8a, 8b, 10b, 10a | 9 | 7, 10, 8, 11 | 11, 8a, 8b |
| 9Hb | 1.79 | 9a, 8b, 8a, 10b | 9 | 7, 10, 8, 11 | 11, 8a, 10a |
| 10Ha | 2.11 | 11, 12, 10b, 9a | 10 | 9, 8, 12, 11 | 12, 9b |
| 10Hb | 2.15 | 11, 12, 10a, 9b, 9a | 10 | 9, 8, 12, 11 | 12 |
| 11H | 5.89 | 12, 6b, 10a, 10b | 11 | 9, 10, 18, 12 | 13, 9a, 9b, 8a, 8b |
| 12H | 6.71 | 11, 10a, 10b | 12 | 18, 11, 13, 17, 10, 9 | 4b, 10a, 10b, 5b, 19 |
| 13H | 6.75 | 15 | 13 | 12, 14, 1, 15, 17 | 11 |
| 15H | 6.65 | 13 | 15 | 17, 14, 16, 1, 13 | |
| 19H3 | 1.29 | 3 | 19 | 4, 5, 3, 17 | 12, 5b |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, Y.; Drouin, P.; Lepp, D.; Li, X.-Z.; Zhu, H.; Castex, M.; Zhou, T. A Novel Microbial Zearalenone Transformation through Phosphorylation. Toxins 2021, 13, 294. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins13050294
Zhu Y, Drouin P, Lepp D, Li X-Z, Zhu H, Castex M, Zhou T. A Novel Microbial Zearalenone Transformation through Phosphorylation. Toxins. 2021; 13(5):294. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins13050294
Chicago/Turabian StyleZhu, Yan, Pascal Drouin, Dion Lepp, Xiu-Zhen Li, Honghui Zhu, Mathieu Castex, and Ting Zhou. 2021. "A Novel Microbial Zearalenone Transformation through Phosphorylation" Toxins 13, no. 5: 294. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins13050294





