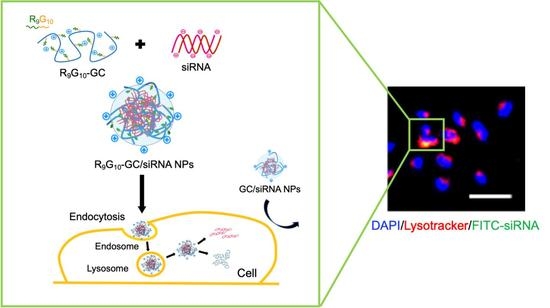
In Vitro Cellular Uptake and Transfection of Oligoarginine-Conjugated Glycol Chitosan/siRNA Nanoparticles
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Synthesis of Peptide-Modified Glycol Chitosan
2.3. Characterization of Peptide-Modified Glycol Chitosan
2.4. Preparation of Peptide-Modified Glycol Chitosan/siRNA Nanoparticles
2.5. Electrophoretic Mobility Shift Assay
2.6. Particle Size and Zeta Potential
2.7. Cell Culture and Cytotoxicity Assay
2.8. Cellular Uptake
2.9. Gene Silencing
2.10. Statistical Analysis
3. Results and Discussion
3.1. Characterization of Peptide-Modified Glycol Chitosan
3.2. Interactions between siRNA and Peptide-Modified Glycol Chitosan
3.3. Size and Surface Charge of Peptide-Modified Glycol Chitosan/siRNA Nanoparticles
3.4. Cytotoxicity
3.5. Cellular Uptake
3.6. Gene Silencing
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shankar, P.; Manjunath, N.; Lieberman, J. The prospect of silencing disease using RNA interference. JAMA 2005, 293, 1367–1373. [Google Scholar] [CrossRef] [PubMed]
- Pecot, C.V.; Calin, G.A.; Coleman, R.L.; Lopez-Berestein, G.; Sood, A.K. RNA interference in the clinic: Challenges and future directions. Nat. Rev. Cancer 2011, 11, 59–67. [Google Scholar] [CrossRef] [Green Version]
- Setten, R.L.; Rossi, J.J.; Han, S. The current state and future directions of RNAi-based therapeutics. Nat. Rev. Drug Discov. 2019, 18, 421–446. [Google Scholar] [CrossRef]
- Conde, J.; Ambrosone, A.; Hernandez, Y.; Tian, F.; McCully, M.; Berry, C.C.; Baptista, P.; Tortiglione, C.; de la Fuente, J.M. 15 years on siRNA delivery: Beyond the state-of-the-art on inorganic nanoparticles for RNAi therapeutics. Nano Today 2015, 10, 421–450. [Google Scholar] [CrossRef] [Green Version]
- Hattab, D.; Bakhtiar, A. Bioengineered siRNA-Based Nanoplatforms Targeting molecular signaling pathways for the treatment of triple negative breast cancer: Preclinical and clinical advancements. Pharmaceutics 2020, 12, 929. [Google Scholar] [CrossRef] [PubMed]
- Fire, A.; Xu, S.Q.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef]
- Plasterk, R.H.A. RNA silencing: The genome’s immune system. Science 2002, 296, 1263–1265. [Google Scholar] [CrossRef] [PubMed]
- Fougerolles, A.; Vornlocher, H.P.; Maraganore, J.; Lieberman, J. Interfering with disease: A progress report on siRNA-based therapeutics. Nat. Rev. Discov. 2008, 6, 443–453. [Google Scholar] [CrossRef]
- Santel, A.; Aleku, M.; Keil, O.; Endruschat, J.; Esche, V.; Durieux, B.; Löffler, K.; Fechtner, M.; Röhl, T.; Fisch, G.; et al. RNA interference in the mouse vascular endothelium by systemic administration of siRNA-lipoplexes for cancer therapy. Gene Ther. 2006, 13, 1360–1370. [Google Scholar] [CrossRef]
- Kumar, P.; Ban, H.S.; Kim, S.S.; Wu, H.Q.; Pearson, T.; Greiner, D.L.; Laouar, A.; Yao, J.; Haridas, V.; Habiro, K.; et al. T cell-specific siRNA delivery suppresses HIV-1 infection in humanized mice. Cell 2008, 134, 577–586. [Google Scholar] [CrossRef] [Green Version]
- Weng, Y.; Xiao, H.; Zhang, J.; Liang, X.-J.; Huang, Y. RNAi therapeutic and its innovative biotechnological evolution. Biotechnol. Adv. 2019, 37, 801–825. [Google Scholar] [CrossRef] [PubMed]
- Schaffert, D.; Wagner, E. Gene therapy progress and prospects: Synthetic polymer-based systems. Gene Ther. 2008, 15, 1131–1138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davis, M.E.; Zuckerman, J.E.; Choi, C.H.J.; Seligson, D.; Tolcher, A.; Alabi, C.A.; Yen, Y.; Heidel, J.D.; Ribas, A. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature 2010, 464, 1067–1070. [Google Scholar] [CrossRef]
- Ozcan, G.; Ozpolat, B.; Coleman, R.L.; Sood, A.K.; Lopez-Berestein, G. Preclinical and clinical development of siRNA-based therapeutics. Adv. Drug Deliv. Rev. 2015, 87, 108–119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hattab, D.; Gazzali, A.M.; Bakhtiar, A. Clinical advances of siRNA-based nanotherapeutics for cancer treatment. Pharmaceutics 2021, 13, 1009. [Google Scholar] [CrossRef]
- Zhang, M.M.; Bahal, R.; Rasmussen, T.P.; Manautou, J.E.; Zhong, X. The growth of siRNA-based therapeutics: Updated clinical studies. Biochem. Pharmacol. 2021, 189, 114432. [Google Scholar] [CrossRef] [PubMed]
- Lundstrom, K. Latest development in viral vectors for gene therapy. Trends Biotechnol. 2003, 21, 117–122. [Google Scholar] [CrossRef]
- Li, S.-D.; Huang, L. Non-viral is superior to viral gene delivery. J. Control. Release 2007, 123, 181–183. [Google Scholar] [CrossRef]
- Park, T.G.; Jeong, J.H.; Kim, S.W. Current status of polymeric gene delivery systems. Adv. Drug Deliv. Rev. 2006, 58, 467–486. [Google Scholar] [CrossRef]
- Pack, D.W.; Hoffman, A.S.; Pun, S.; Stayton, P.S. Design and development of polymers for gene delivery. Nat. Rev. Drug Discov. 2005, 4, 581–593. [Google Scholar] [CrossRef]
- Whitehead, K.A.; Langer, R.; Anderson, D.G. Knocking down barriers: Advances in siRNA delivery. Nat. Rev. Drug Discov. 2009, 8, 129–138. [Google Scholar] [CrossRef]
- Miyata, K.; Kakizawa, Y.; Nishiyama, N.; Harada, A.; Yamasaki, Y.; Koyama, H.; Kataoka, K. Block catiomer polyplexes with regulated densities of charge and disulfide cross-linking directed to enhance gene expression. J. Am. Chem. Soc. 2004, 126, 2355–2361. [Google Scholar] [CrossRef] [PubMed]
- Truong, N.P.; Gu, W.; Prasadam, I.; Jia, Z.; Crawford, R.; Xiao, Y.; Monteiro, M.J. An influenza virus-inspired polymer system for the timed release of siRNA. Nature Comm. 2013, 4, 1902. [Google Scholar] [CrossRef] [Green Version]
- Hartono, S.B.; Phuoc, N.T.; Yu, M.; Jia, Z.; Monteiro, M.J.; Qiao, S.; Yu, C. Functionalized large pore mesoporous silica nanoparticles for gene delivery featuring controlled release and co-delivery. J. Mater. Chem. B 2014, 2, 718–726. [Google Scholar] [CrossRef] [PubMed]
- Muzzarelli, R.A.A.; Boudrant, J.; Meyer, D.; Manno, N.; DeMarchis, M.; Paoletti, M.G. Current views on fungal chitin/chitosan, human chitinases, food preservation, glucans, pectins and inulin: A tribute to Henri Braconnot, precursor of the carbohydrate polymers science, on the chitin bicentennial. Carbohydr. Polym. 2012, 87, 995–1012. [Google Scholar] [CrossRef]
- Zargar, V.; Asghari, M.; Dashti, A. A review on chitin and chitosan polymers: Structure, chemistry, solubility, derivatives, and applications. ChemBioEng Rev. 2015, 2, 204–226. [Google Scholar] [CrossRef]
- Cao, Y.; Tan, Y.F.; Wong, Y.S.; Liew, M.W.J.; Venkatraman, S. Recent advances in chitosan-based carriers for gene delivery. Marine Drugs 2019, 17, 381. [Google Scholar] [CrossRef] [Green Version]
- Chuan, D.; Jin, T.; Fan, R.; Zhou, L.; Guo, G. Chitosan for gene delivery: Methods for improvement and applications. Adv. Colloid Interface Sci. 2019, 268, 25–38. [Google Scholar] [CrossRef]
- Bravo-Anaya, L.M.; Fernandez-Solis, K.G.; Rosselgong, J.; Nano-Rodriguez, J.L.E.; Carvajal, F.; Rinaudo, M. Chitosan-DNA polyelectrolyte complex: Influence of chitosan characteristics and mechanism of complex formation. Int. J. Biol. Macromol. 2019, 126, 1037–1049. [Google Scholar] [CrossRef] [PubMed]
- Azzam, M.; El Safy, S.; Abdelgelil, S.A.; Weiskirchen, R.; Asimakopoulou, A.; de Lorenzi, F.; Lammers, T.; Mansour, S.; Tammam, S. Targeting activated hepatic stellate cells using collagen-binding chitosan nanoparticles for siRNA delivery to fibrotic livers. Pharmaceutics 2021, 12, 590. [Google Scholar] [CrossRef] [PubMed]
- Ashrafizadeh, M.; Delfi, M.; Hashemi, F.; Zabolian, A.; Saleki, H.; Bagherian, M.; Azami, N.; Farahani, M.V.; Farahani, S.O.; Farahani, S.; et al. Biomedical application of chitosan-based nanoscale delivery systems: Potential usefulness in siRNA delivery for cancer therapy. Carbohydr. Polym. 2021, 260, 117809. [Google Scholar] [CrossRef] [PubMed]
- Alameh, M.; Lavertu, M.; Tran-Khanh, N.; Chang, C.-Y.; Lesage, F.; Bail, M.; Darras, V.; Chetvrier, A.; Buschmann, M.D. siRNA delivery with chitosan: Influence of chitosan molecular weight, degree of deacetylation, and amine to phosphate ratio on in vitro silencing efficiency, hemocompatibility, biodistribution, and in vivo efficacy. Biomacromolecules 2018, 19, 112–131. [Google Scholar] [CrossRef] [PubMed]
- Yoon, H.Y.; Son, S.; Lee, S.J.; You, D.G.; Yhee, J.Y.; Park, J.H.; Swierczewska, M.; Lee, S.; Kwon, I.C.; Kim, S.H.; et al. Glycol chitosan nanoparticles as specialized cancer therapeutic vehicles: Sequential delivery of doxorubicin and Bcl-2 siRNA. Sci. Rep. 2014, 4, 6878. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Y.; Han, S.; Liang, Z.; Zhao, M.; Liu, G.; Wu, J. Progress in arginine-based gene delivery systems. J. Mater. Chem. B 2020, 8, 5564–5577. [Google Scholar] [CrossRef]
- Kim, S.W.; Kim, N.Y.; Bin Choi, Y.; Park, S.H.; Yang, J.M.; Shin, S. RNA interference in vitro and in vivo using an arginine peptide/siRNA complex system. J. Control. Release 2010, 143, 335–343. [Google Scholar] [CrossRef]
- Zhao, X.; Li, Z.; Liu, W.; Lam, W.; Sun, P.; Kao, R.Y.T.; Luk, K.D.K.; Lu, W.W. Octaarginine-modified chitosan as a nonviral gene delivery vector: Properties and in vitro transfection efficiency. J. Nanoparticle Res. 2011, 13, 693–702. [Google Scholar] [CrossRef]
- Futaki, S.; Suzuki, T.; Ohashi, W.; Yagami, T.; Tanaka, S.; Ueda, K.; Sugiura, Y. Arginine-rich peptides. An abundant source of membrane-permeable peptides having potential as carriers for intracellular protein delivery. J. Biol. Chem. 2001, 276, 5836–5840. [Google Scholar] [CrossRef] [Green Version]
- Saw, P.E.; Ko, Y.T.; Jon, S. Efficient liposomal nanocarrier-mediated oligodeoxynucleotide delivery involving dual use of a cell-penetrating peptide as a packaging and intracellular delivery agent. Macromol. Rapid Commun. 2010, 31, 1155–1162. [Google Scholar] [CrossRef]
- Park, S.; Jeong, E.J.; Lee, J.; Rhim, T.; Lee, S.K.; Lee, K.Y. Preparation and characterization of nonaarginine-modified chitosan nanoparticles for siRNA delivery. Carbohydr. Polym. 2013, 92, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.W.; Park, Y.-J.; Lee, S.J.; Lee, S.K.; Lee, K.Y. The effect of spacer arm length of an adhesion ligand coupled to an alginate gel on the control of fibroblast phenotype. Biomaterials 2010, 31, 5545–5551. [Google Scholar] [CrossRef] [PubMed]
- Hermanson, G.T. Bioconjugate Techniques; Academic Press: San Diego, CA, USA, 2013; pp. 259–266. [Google Scholar]
- Dumont, V.C.; Mansur, H.S.; Mansur, A.A.P.; Carvalho, S.M.; Capanema, N.S.V.; Barrioni, B.R. Glycol chitosan/nanohydroxyapatite biocomposites for potential bone tissue engineering and regenerative medicine. Int. J. Biol. Macromol. 2016, 93, 1465–1478. [Google Scholar] [CrossRef]
- Barreira, S.V.; Silva, F. Surface modification chemistry based on the electrostatic adsorption of poly-L-arginine onto alkanethiol modified gold surfaces. Langmuir 2003, 19, 10324–10331. [Google Scholar] [CrossRef]
- Kim, T.-I.; Ou, M.; Lee, M.; Kim, S.W. Arginine-grafted bioreducible poly (disulfide amine) for gene delivery systems. Biomaterials 2009, 30, 658–664. [Google Scholar] [CrossRef] [Green Version]
- Jeong, E.J.; Choi, M.; Lee, J.; Rhim, T.; Lee, K.Y. The spacer arm length in cell-penetrating peptides influences chitosan/siRNA nanoparticle delivery for pulmonary inflammation treatment. Nanoscale 2015, 7, 20095–20104. [Google Scholar] [CrossRef] [PubMed]
- Vu, M.N.; Kelly, H.G.; Wheatley, A.K.; Peng, S.; Pilkington, E.H.; Veldhuis, N.A.; Davis, T.P.; Kent, S.J.; Truong, N.P. Cellular interactions of piposomes and PISA nanoparticles during human blood flow in a microvascular network. Small 2020, 16, 2002861. [Google Scholar] [CrossRef]
- He, C.; Hu, Y.; Yin, L.; Tang, C.; Yin, C. Effects of particle size and surface charge on cellular uptake and biodistribution of polymeric nanoparticles. Biomaterials 2010, 31, 3657–3666. [Google Scholar] [CrossRef] [PubMed]
- Truong, N.P.; Jia, Z.; Burgess, M.; Payne, L.; McMillan, N.A.J.; Monteiro, M.J. Self-catalyzed degradable cationic polymer for release of DNA. Biomacromolecules 2011, 12, 3540–3548. [Google Scholar] [CrossRef] [PubMed]








| Sample | Theoretical DS a | Actual DS | Conjugation Efficiency (%) |
|---|---|---|---|
| R9G10-GC 0.075 | 0.075 | 0.071 | 94.6 |
| R9G10-GC 0.6 | 0.6 | 0.544 | 90.7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jeong, E.-J.; Lee, J.; Kim, H.-S.; Lee, K.-Y. In Vitro Cellular Uptake and Transfection of Oligoarginine-Conjugated Glycol Chitosan/siRNA Nanoparticles. Polymers 2021, 13, 4219. https://0-doi-org.brum.beds.ac.uk/10.3390/polym13234219
Jeong E-J, Lee J, Kim H-S, Lee K-Y. In Vitro Cellular Uptake and Transfection of Oligoarginine-Conjugated Glycol Chitosan/siRNA Nanoparticles. Polymers. 2021; 13(23):4219. https://0-doi-org.brum.beds.ac.uk/10.3390/polym13234219
Chicago/Turabian StyleJeong, Eun-Ju, Jangwook Lee, Hyun-Seung Kim, and Kuen-Yong Lee. 2021. "In Vitro Cellular Uptake and Transfection of Oligoarginine-Conjugated Glycol Chitosan/siRNA Nanoparticles" Polymers 13, no. 23: 4219. https://0-doi-org.brum.beds.ac.uk/10.3390/polym13234219