Biomass Prediction of Heterogeneous Temperate Grasslands Using an SfM Approach Based on UAV Imaging
Abstract
:1. Introduction
2. Materials and Methods
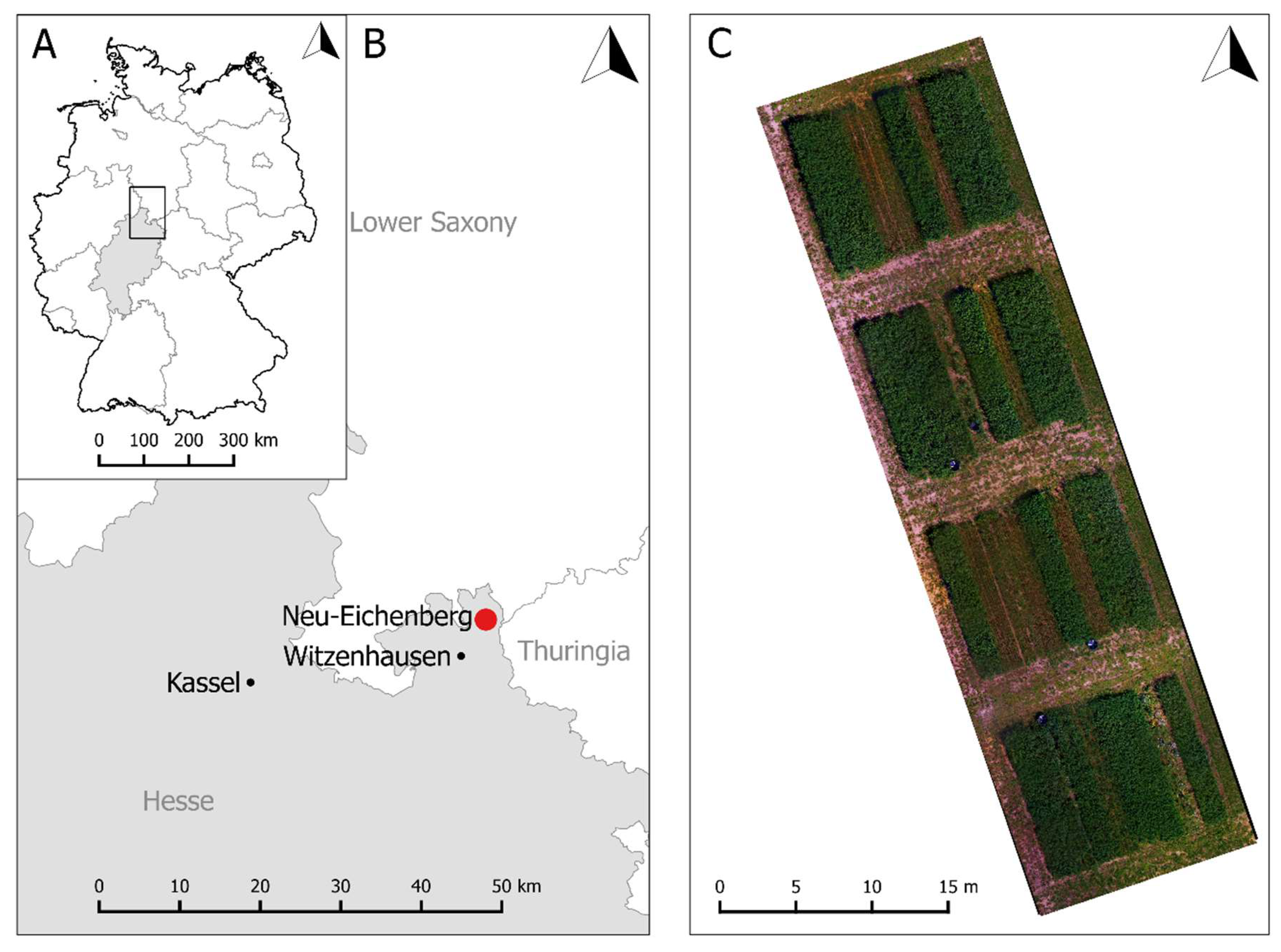
2.1. Experimental Site and Design
2.2. RGB Remote Sensing and Data Acquisition
2.3. Data Processing and Analysis
2.4. Statistical Analysis
3. Results
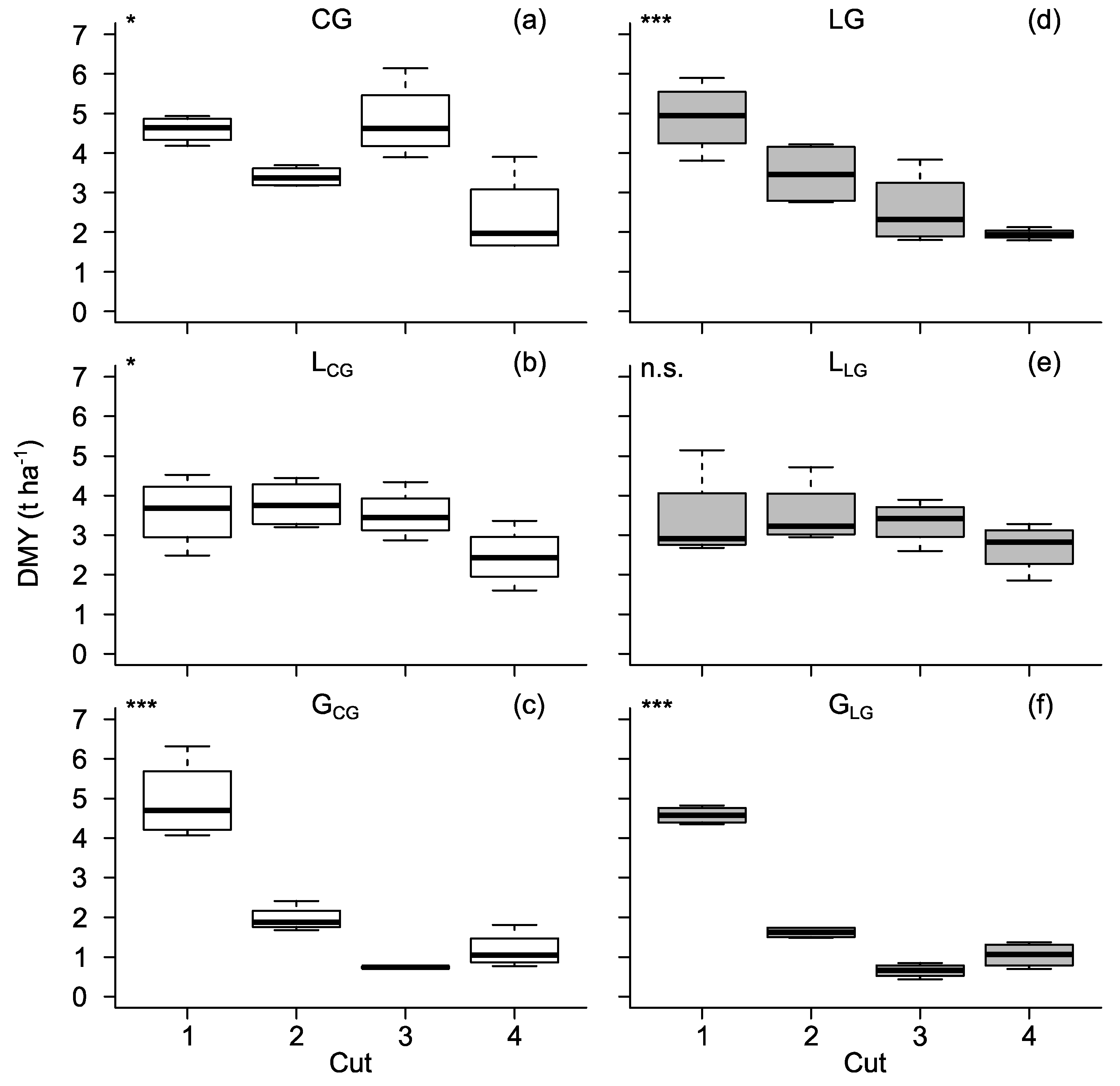
3.1. Dry Matter Yield
3.2. Canopy Height
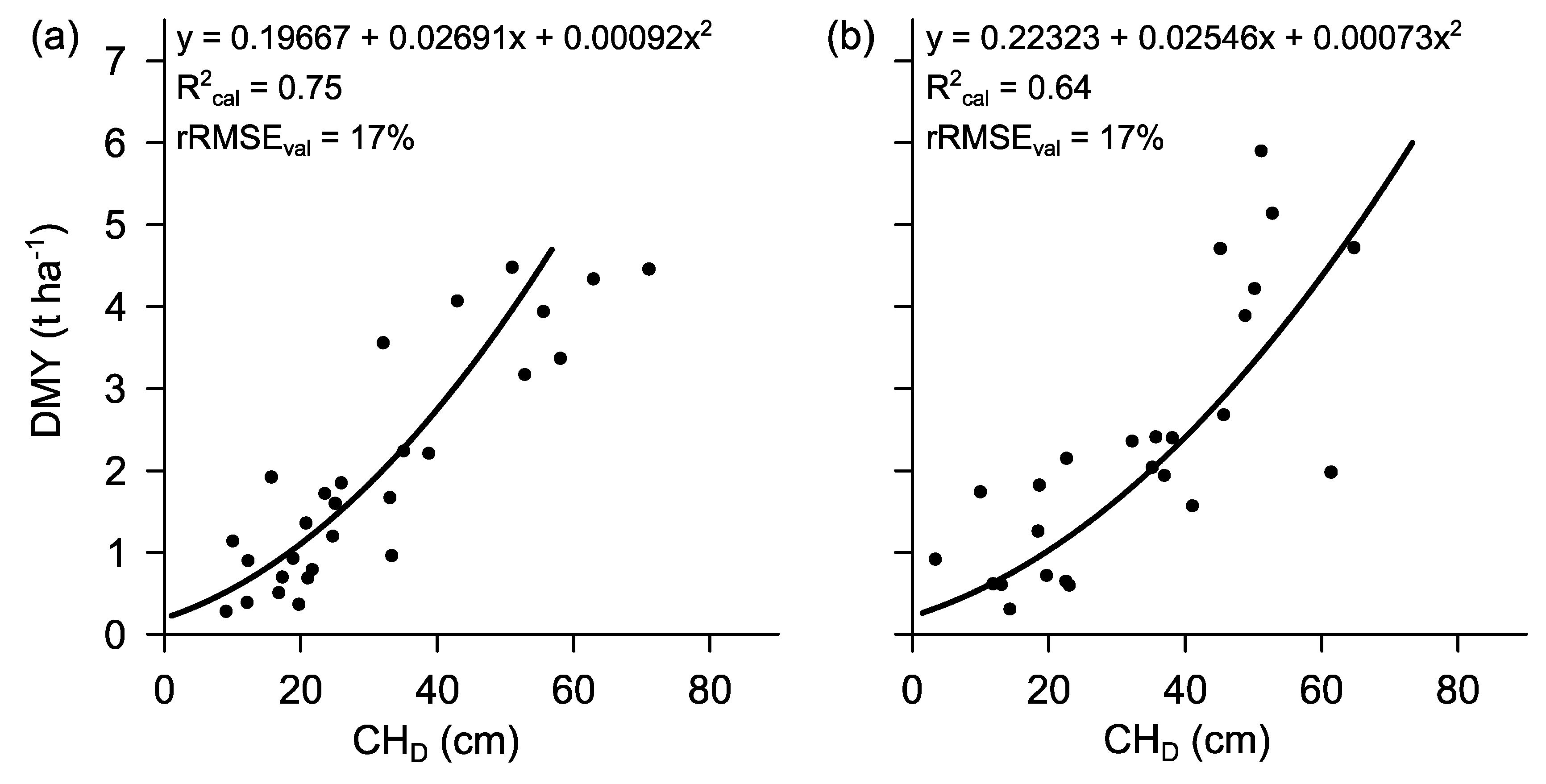
3.3. Prediction Models
4. Discussion
5. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
Appendix A
| Cut Date | DMY (t ha−1) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1st harvest | 1st sub-sample | 2nd sub-sample | 2nd harvest | 3rd sub-sample | 3rd harvest | 4th sub-sample | 5th sub-sample | 6th sub-sample | 4th harvest | |
| 17 May 2017 | 2 June 2017 | 13 June 2017 | 26 June 2017 | 11 July 2017 | 8 August 2017 | 23 August 2017 | 5 September 2017 | 20 September 2017 | 9 October 2017 | |
| Treatment | ||||||||||
| CG | 4.60 | 0.78 | 2.29 | 3.40 | 0.73 | 4.82 | 0.44 | 1.15 | 1.79 | 2.37 |
| LG | 4.90 | 0.60 | 2.21 | 3.47 | 0.67 | 2.57 | 0.78 | 1.70 | 1.87 | 1.95 |
| LCG | 4.95 | 0.35 | 1.49 | 1.97 | 0.29 | 0.74 | 0.33 | 0.87 | 1.35 | 1.17 |
| LLG | 3.59 | 1.12 | 2.59 | 3.79 | 1.03 | 3.53 | 0.78 | 2.06 | 2.93 | 2.46 |
| GCG | 4.58 | 0.57 | 1.59 | 1.62 | 0.21 | 0.66 | 0.41 | 0.86 | 1.08 | 1.05 |
| GLG | 3.41 | 0.76 | 1.97 | 3.53 | 0.95 | 3.33 | 0.52 | 1.81 | 1.81 | 2.70 |
References
- Sanderson, M.A.; Rotz, C.A.; Fultz, S.W.; Rayburn, E.B. Estimating Forage Mass with a Commercial Capacitance Meter, Rising Plate Meter, and Pasture Ruler. Agron. J. 2001, 93, 1281. [Google Scholar] [CrossRef]
- Schellberg, J.; Hill, M.J.; Gerhards, R.; Rothmund, M.; Braun, M. Precision agriculture on grassland: Applications, perspectives and constraints. Eur. J. Agron. 2008, 29, 59–71. [Google Scholar] [CrossRef]
- Wachendorf, M.; Fricke, T.; Möckel, T. Remote sensing as a tool to assess botanical composition, structure, quantity and quality of temperate grasslands. Grass Forage Sci. 2017, 35, 201. [Google Scholar] [CrossRef]
- Catchpole, E.R.; Wheeler, C.J. Estimating plant biomass: A review of techniques. Aust. J. Ecol. 1992, 121–131. [Google Scholar] [CrossRef]
- Fricke, T.; Wachendorf, M. Combining ultrasonic sward height and spectral signatures to assess the biomass of legume–grass swards. Comput. Electron. Agric. 2013, 99, 236–247. [Google Scholar] [CrossRef]
- Holman, F.; Riche, A.; Michalski, A.; Castle, M.; Wooster, M.; Hawkesford, M. High Throughput Field Phenotyping of Wheat Plant Height and Growth Rate in Field Plot Trials Using UAV Based Remote Sensing. Remote Sens. 2016, 8, 1031. [Google Scholar] [CrossRef]
- Hakl, J.; Hrevušová, Z.; Hejcman, M.; Fuksa, P. The use of a rising plate meter to evaluate lucerne (Medicago sativa L.) height as an important agronomic trait enabling yield estimation. Grass Forage Sci. 2012, 67, 589–596. [Google Scholar] [CrossRef]
- Ondřej, C.; Josef, H.; Michal, H.; Pavel, C. The use of compressed height to estimate the yield of a differently fertilized meadow. Plant Soil Environ. 2018, 64, 76–81. [Google Scholar] [CrossRef] [Green Version]
- Harmoney, K.R.; Moore, K.J.; George, J.R.; Brummer, E.C.; Russell, J.R. Determination of Pasture Biomass Using Four Indirect Methods. Agron. J. 1997, 89, 665. [Google Scholar] [CrossRef]
- Künnemeyer, R.; Schaare, P.N.; Hanna, M.M. A simple reflectometer for on-farm pasture assessment. Comput. Electron. Agric. 2001, 31, 125–136. [Google Scholar] [CrossRef]
- Lu, D. The potential and challenge of remote sensing-based biomass estimation. Int. J. Remote Sens. 2006, 27, 1297–1328. [Google Scholar] [CrossRef]
- Moeckel, T.; Safari, H.; Reddersen, B.; Fricke, T.; Wachendorf, M. Fusion of Ultrasonic and Spectral Sensor Data for Improving the Estimation of Biomass in Grasslands with Heterogeneous Sward Structure. Remote Sens. 2017, 9, 98. [Google Scholar] [CrossRef]
- Wallace, L.; Hillman, S.; Reinke, K.; Hally, B.; Kriticos, D. Non-destructive estimation of above-ground surface and near-surface biomass using 3D terrestrial remote sensing techniques. Methods Ecol. Evol. 2017, 257, 1684. [Google Scholar] [CrossRef]
- Forsmoo, J.; Anderson, K.; Macleod, C.J.A.; Wilkinson, M.E.; Brazier, R.; Smit, I. Drone-based structure-from-motion photogrammetry captures grassland sward height variability. J. Appl. Ecol. 2018, 94, 237. [Google Scholar] [CrossRef]
- Dittmann, S.; Thiessen, E.; Hartung, E. Applicability of different non-invasive methods for tree mass estimation: A review. For. Ecol. Manag. 2017, 398, 208–215. [Google Scholar] [CrossRef]
- Schirrmann, M.; Giebel, A.; Gleiniger, F.; Pflanz, M.; Lentschke, J.; Dammer, K.-H. Monitoring Agronomic Parameters of Winter Wheat Crops with Low-Cost UAV Imagery. Remote Sens. 2016, 8, 706. [Google Scholar] [CrossRef]
- Bendig, J.; Bolten, A.; Bennertz, S.; Broscheit, J.; Eichfuss, S.; Bareth, G. Estimating Biomass of Barley Using Crop Surface Models (CSMs) Derived from UAV-Based RGB Imaging. Remote Sens. 2014, 6, 10395–10412. [Google Scholar] [CrossRef] [Green Version]
- Geipel, J.; Link, J.; Claupein, W. Combined Spectral and Spatial Modeling of Corn Yield Based on Aerial Images and Crop Surface Models Acquired with an Unmanned Aircraft System. Remote Sens. 2014, 6, 10335–10355. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Niu, Z.; Chen, H.; Li, D.; Wu, M.; Zhao, W. Remote estimation of canopy height and aboveground biomass of maize using high-resolution stereo images from a low-cost unmanned aerial vehicle system. Ecol. Indic. 2016, 67, 637–648. [Google Scholar] [CrossRef]
- Moeckel, T.; Dayananda, S.; Nidamanuri, R.; Nautiyal, S.; Hanumaiah, N.; Buerkert, A.; Wachendorf, M. Estimation of Vegetable Crop Parameter by Multi-temporal UAV-Borne Images. Remote Sens. 2018, 10, 805. [Google Scholar] [CrossRef]
- Ergon, Å.; Kirwan, L.; Bleken, M.A.; Skjelvåg, A.O.; Collins, R.P.; Rognli, O.A. Species interactions in a grassland mixture under low nitrogen fertilization and two cutting frequencies: 1. dry-matter yield and dynamics of species composition. Grass Forage Sci. 2016, 71, 667–682. [Google Scholar] [CrossRef] [Green Version]
- Elgersma, A.; Søegaard, K. Changes in nutritive value and herbage yield during extended growth intervals in grass-legume mixtures: Effects of species, maturity at harvest, and relationships between productivity and components of feed quality. Grass Forage Sci. 2018, 73, 78–93. [Google Scholar] [CrossRef]
- Cooper, S.; Roy, D.; Schaaf, C.; Paynter, I. Examination of the Potential of Terrestrial Laser Scanning and Structure-from-Motion Photogrammetry for Rapid Nondestructive Field Measurement of Grass Biomass. Remote Sens. 2017, 9, 531. [Google Scholar] [CrossRef]
- Van Iersel, W.; Straatsma, M.; Addink, E.; Middelkoop, H. Monitoring height and greenness of non-woody floodplain vegetation with UAV time series. ISPRS J. Photogramm. Remote Sens. 2018, 141, 112–123. [Google Scholar] [CrossRef]
- Viljanen, N.; Honkavaara, E.; Näsi, R.; Hakala, T.; Niemeläinen, O.; Kaivosoja, J. A Novel Machine Learning Method for Estimating Biomass of Grass Swards Using a Photogrammetric Canopy Height Model, Images and Vegetation Indices Captured by a Drone. Agriculture 2018, 8, 70. [Google Scholar] [CrossRef]
- Heady, H.F. The Measurement and Value of Plant Height in the Study of Herbaceous Vegetation. Ecology 1957, 38, 313–320. [Google Scholar] [CrossRef]
- Cohen, W.B.; Maiersperger, T.K.; Gower, S.T.; Turner, D.P. An improved strategy for regression of biophysical variables and Landsat ETM+ data. Remote Sens. Environ. 2003, 84, 561–571. [Google Scholar] [CrossRef] [Green Version]
- Willmott, C.J. On the validation of models. Phys. Geogr. 1981, 184–194. [Google Scholar] [CrossRef]
- Aasen, H.; Burkart, A.; Bolten, A.; Bareth, G. Generating 3D hyperspectral information with lightweight UAV snapshot cameras for vegetation monitoring: From camera calibration to quality assurance. ISPRS J. Photogramm. Remote Sens. 2015, 108, 245–259. [Google Scholar] [CrossRef]
- Cunliffe, A.M.; Brazier, R.E.; Anderson, K. Ultra-fine grain landscape-scale quantification of dryland vegetation structure with drone-acquired structure-from-motion photogrammetry. Remote Sens. Environ. 2016, 183, 129–143. [Google Scholar] [CrossRef] [Green Version]
- Malambo, L.; Popescu, S.C.; Murray, S.C.; Putman, E.; Pugh, N.A.; Horne, D.W.; Richardson, G.; Sheridan, R.; Rooney, W.L.; Avant, R.; et al. Multitemporal field-based plant height estimation using 3D point clouds generated from small unmanned aerial systems high-resolution imagery. Int. J. Appl. Earth Obs. Geoinf. 2018, 64, 31–42. [Google Scholar] [CrossRef]
- Roth, L.; Streit, B. Predicting cover crop biomass by lightweight UAS-based RGB and NIR photography: An applied photogrammetric approach. Precis. Agric. 2018, 19, 93–114. [Google Scholar] [CrossRef]
- Bendig, J.; Yu, K.; Aasen, H.; Bolten, A.; Bennertz, S.; Broscheit, J.; Gnyp, M.L.; Bareth, G. Combining UAV-based plant height from crop surface models, visible, and near infrared vegetation indices for biomass monitoring in barley. Int. J. Appl. Earth Obs. Geoinf. 2015, 39, 79–87. [Google Scholar] [CrossRef]
- Grenzdörffer, G.J. Crop height determination with UAS point clouds. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2014, XL-1, 135–140. [Google Scholar] [CrossRef]
- Gillan, J.K.; Karl, J.W.; Duniway, M.; Elaksher, A. Modeling vegetation heights from high resolution stereo aerial photography: An application for broad-scale rangeland monitoring. J. Environ. Manag. 2014, 144, 226–235. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, K.; Guo, W.; Arai, K.; Takanashi, H.; Kajiya-Kanegae, H.; Kobayashi, M.; Yano, K.; Tokunaga, T.; Fujiwara, T.; Tsutsumi, N.; et al. High-Throughput Phenotyping of Sorghum Plant Height Using an Unmanned Aerial Vehicle and Its Application to Genomic Prediction Modeling. Front. Plant Sci. 2017, 8, 421. [Google Scholar] [CrossRef] [PubMed]
- Schut, A.G.T.; Traore, P.C.S.; Blaes, X.; de By, R.A. Assessing yield and fertilizer response in heterogeneous smallholder fields with UAVs and satellites. Field Crop. Res. 2018, 221, 98–107. [Google Scholar] [CrossRef]
- Ledgard, S.F.; Steele, K.W. Biological nitrogen fixation in mixed legume/grass pastures. Plant Soil 1992, 141, 137–153. [Google Scholar] [CrossRef]


| Treatment | Functional Group | Species | Ratio (%) | |
|---|---|---|---|---|
| Clover-grass mixture | CG | Legumes (L) | Trifolium pratense Trifolium hybridum Trifolium repens | 30 5 5 |
| Grass (G) | Lolium multiflorum | 60 | ||
| Lucerne-grass mixture | LG | L | Medicago sativa Trifolium pratense | 40 10 |
| G | Festuca pratensis Lolium perenne Lolium multiflorum Phleum pratense | 20 15 10 5 | ||
| Pure clover legumes | LCG | L from CG mixture | Trifolium pratense Trifolium hybridum Trifolium repens | 75 12.5 12.5 |
| Pure lucerne and clover legumes | LLG | L from LG mixture | Medicago sativa Trifolium pratense | 80 20 |
| Pure grass sward | GCG | G from CG mixture | Lolium multiflorum | 100 |
| Pure grass sward | GLG | G from LG mixture | Festuca pratensis Lolium perenne Lolium multiflorum Phleum pratense | 40 30 20 10 |
| Treatment | R2 | RMSE (cm) | rRMSE (%) |
|---|---|---|---|
| All | 0.56 (0.70) | 13.39 (10.32) | 17 (13) |
| CG | 0.79 | 10.19 | 13 |
| LG | 0.70 | 11.14 | 16 |
| LCG | 0.84 | 6.08 | 11 |
| LLG | 0.72 | 8.70 | 16 |
| GCG | 0.47 (0.70) | 16.51 (9.17) | 22 (14) |
| GLG | 0.29 (0.57) | 16.91 (9.35) | 24 (17) |
| Treatment | Calibration | Validation | |||||
|---|---|---|---|---|---|---|---|
| ncal | R2cal | nval | R2val | RMSEval (t ha−1) | rRMSEval (%) | d | |
| CHR | |||||||
| All | 180 (174) | 0.62 (0.71) | 53 (51) | 0.64 (0.65) | 0.28 (0.29) | 18 (19) | 0.90 (0.90) |
| CG | 30 | 0.80 | 10 | 0.66 | 0.34 | 19 | 0.90 |
| LG | 30 | 0.71 | 9 | 0.50 | 0.33 | 19 | 0.85 |
| LCG | 30 | 0.68 | 10 | 0.56 | 0.26 | 21 | 0.87 |
| LLG | 30 | 0.77 | 8 | 0.68 | 0.23 | 19 | 0.91 |
| GCG | 30 (27) | 0.64 (0.82) | 8 (7) | 0.42 (0.51) | 0.33 (0.34) | 21 (22) | 0.83 (0.88) |
| GLG | 30 (27) | 0.58 (0.82) | 8 (7) | 0.43 (0.73) | 0.29 (0.20) | 22 (15) | 0.82 (0.94) |
| CHD | |||||||
| All | 180 (174) | 0.69 (0.73) | 53 (51) | 0.72 (0.62) | 0.27 (0.30) | 17 (20) | 0.92 (0.89) |
| CG | 30 | 0.80 | 10 | 0.87 | 0.23 | 13 | 0.96 |
| LG | 30 | 0.63 | 9 | 0.68 | 0.35 | 20 | 0.89 |
| LCG | 30 | 0.81 | 10 | 0.46 | 0.29 | 24 | 0.83 |
| LLG | 30 | 0.62 | 8 | 0.51 | 0.36 | 30 | 0.82 |
| GCG | 30 (27) | 0.68 (0.69) | 8 (7) | 0.54 (0.64) | 0.35 (0.30) | 23 (19) | 0.85 (0.90) |
| GLG | 30 (27) | 0.67 (0.71) | 8 (7) | 0.48 (0.77) | 0.30 (0.20) | 23 (16) | 0.86 (0.94) |
| Treatment | Calibration | Validation | |||||
|---|---|---|---|---|---|---|---|
| ncal | R2cal | nval | R2val | RMSEval (t ha−1) | rRMSEval (%) | d | |
| CHR | |||||||
| Clover-grass (CG, LCG, GCG) Lucerne-grass (LG, LLG, GLG) | 90 | 0.60 | 28 | 0.58 | 0.34 | 22 | 0.87 |
| 90 | 0.65 | 25 | 0.69 | 0.29 | 16 | 0.90 | |
| CHD | |||||||
| Clover-grass (CG, LCG, GCG) Lucerne-grass (LG, LLG, GLG) | 90 | 0.75 | 28 | 0.75 | 0.26 | 17 | 0.93 |
| 90 | 0.64 | 25 | 0.62 | 0.32 | 17 | 0.88 | |
| Treatment | ADMY (t ha−1) | ADMYR (t ha−1) | ADMYD (t ha−1) | |||
|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | |
| All | 11.85 | 2.01 | 12.38 | 1.92 | 11.77 | 2.09 |
| CG | 15.18 | 2.25 | 16.85 | 2.14 | 15.71 | 2.62 |
| LG | 12.88 | 2.37 | 13.73 | 2.17 | 12.30 | 1.76 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grüner, E.; Astor, T.; Wachendorf, M. Biomass Prediction of Heterogeneous Temperate Grasslands Using an SfM Approach Based on UAV Imaging. Agronomy 2019, 9, 54. https://0-doi-org.brum.beds.ac.uk/10.3390/agronomy9020054
Grüner E, Astor T, Wachendorf M. Biomass Prediction of Heterogeneous Temperate Grasslands Using an SfM Approach Based on UAV Imaging. Agronomy. 2019; 9(2):54. https://0-doi-org.brum.beds.ac.uk/10.3390/agronomy9020054
Chicago/Turabian StyleGrüner, Esther, Thomas Astor, and Michael Wachendorf. 2019. "Biomass Prediction of Heterogeneous Temperate Grasslands Using an SfM Approach Based on UAV Imaging" Agronomy 9, no. 2: 54. https://0-doi-org.brum.beds.ac.uk/10.3390/agronomy9020054





