Functional Genomics Analysis to Disentangle the Role of Genetic Variants in Major Depression
Abstract
:1. Introduction
2. Materials and Methods
2.1. MD GWAS Dataset and LD expansion
2.2. GVs Annotation: VEP, CADD and ENCODE
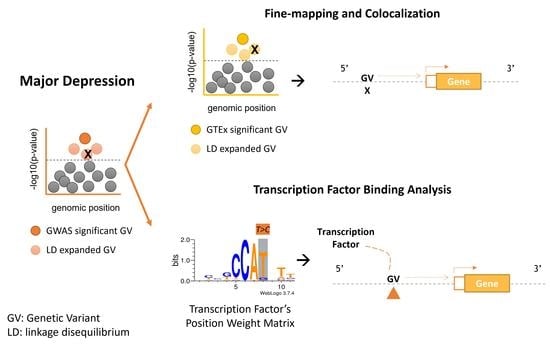
2.3. Fine-Mapping and Colocalization of GWAS and cis-eQTLs
2.4. TF Binding Analysis with RSAT Variation Tools
2.5. Identification of TF Active Regions with ChromHMM
2.6. Retrieval of Regulation Evidence
2.7. pGenes, eGenes, and GVs Characterization
3. Results
3.1. Major Depression Associated Genetic Variants Lie in Non-Coding Regions of the Genome
3.2. Major Depression Causal Genetic Variants Regulate the Expression of Genes in Cis
3.3. MD Associated GVs Affect the TFBS in Regulatory Regions of Genes Relevant for the Disease
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CADD | Combined Annotation Dependent Depletion |
| CLPP | colocalization posterior probability |
| eGenes | genes regulated by eQTLs |
| ENCODE | Encyclopedia of DNA Elements |
| eQTLs | expression quantitative trait loci |
| GO | Gene Ontology |
| GV | genetic variant |
| GTEx | Genotype-Tissue Expression |
| GWAS | genome-wide association studies |
| LD | linkage disequilibrium |
| MD | major depression |
| pGenes | proximal genes |
| PICS | Probabilistic Identification of Causal SNPs |
| PSSM | Position-specific scoring matrix or position weight matrix |
| REAC | Reactome |
| RSAT | Regulatory Sequence Analysis Tools |
| TF | transcription factor |
| TPM | transcripts per million |
| TFBS | transcription factor binding site |
| VEP | variant effect predictor |
References
- World Health Organization: Depression. Available online: https://www.who.int/news-room/fact-sheets/detail/depression (accessed on 21 December 2021).
- Preskorn, S.H. Drug Development in Psychiatry: The Long and Winding Road from Chance Discovery to Rational Development. In Handbook of Experimental Pharmacology; Springer: Berlin/Heidelberg, Germany, 2018; Volume 250, pp. 307–324. [Google Scholar] [CrossRef]
- Howard, D.M.; Adams, M.J.; Clarke, T.-K.; Hafferty, J.D.; Gibson, J.; Shirali, M.; Coleman, J.R.I.; Hagenaars, S.P.; Ward, J.; Wigmore, E.M.; et al. Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat. Neurosci. 2019, 22, 343–352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wray, N.R.; Ripke, S.; Mattheisen, M.; Trzaskowski, M.; Byrne, E.M.; Abdellaoui, A.; Adams, M.J.; Agerbo, E.; Air, T.M.; Andlauer, T.M.F.; et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat. Genet. 2018, 50, 668–681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cano-Gamez, E.; Trynka, G. From GWAS to Function: Using Functional Genomics to Identify the Mechanisms Underlying Complex Diseases. Front. Genet. 2020, 11, 424. [Google Scholar] [CrossRef] [PubMed]
- Umans, B.D.; Battle, A.; Gilad, Y. Where Are the Disease-Associated eQTLs? Trends Genet. 2020, 37, 109–124. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Miao, Y.-R.; Jia, L.-H.; Yu, Q.-Y.; Zhang, Q.; Guo, A.-Y. AnimalTFDB 3.0: A comprehensive resource for annotation and prediction of animal transcription factors. Nucleic Acids Res. 2018, 47, D33–D38. [Google Scholar] [CrossRef] [PubMed]
- Perdomo-Sabogal, Á.; Nowick, K. Genetic Variation in Human Gene Regulatory Factors Uncovers Regulatory Roles in Local Adaptation and Disease. Genome Biol. Evol. 2019, 11, 2178–2193. [Google Scholar] [CrossRef]
- Buniello, A.; MacArthur, J.A.L.; Cerezo, M.; Harris, L.W.; Hayhurst, J.; Malangone, C.; McMahon, A.; Morales, J.; Mountjoy, E.; Sollis, E.; et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019, 47, D1005–D1012. [Google Scholar] [CrossRef] [Green Version]
- Taylor, K.E.; Ansel, K.M.; Marson, A.; Criswell, L.A.; Farh, K.K.-H. PICS2: Next-generation fine mapping via probabilistic identification of causal SNPs. Bioinformatics 2021, 37, 3004–3007. [Google Scholar] [CrossRef]
- McLaren, W.; Gil, L.; Hunt, S.E.; Riat, H.S.; Ritchie, G.R.S.; Thormann, A.; Flicek, P.; Cunningham, F. The Ensembl Variant Effect Predictor. Genome Biol. 2016, 17, 122. [Google Scholar] [CrossRef] [Green Version]
- Rentzsch, P.; Witten, D.; Cooper, G.M.; Shendure, J.; Kircher, M. CADD: Predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019, 47, D886–D894. [Google Scholar] [CrossRef]
- The ENCODE Project Consortium. An Integrated Encyclopedia of DNA Elements in the Human Genome. Nature 2012, 489, 57–74. [Google Scholar] [CrossRef] [PubMed]
- Oscanoa, J.; Sivapalan, L.; Gadaleta, E.; Ullah, A.Z.D.; Lemoine, N.R.; Chelala, C. SNPnexus: A web server for functional annotation of human genome sequence variation (2020 update). Nucleic Acids Res. 2020, 48, W185–W192. [Google Scholar] [CrossRef] [PubMed]
- Ghoussaini, M.; Mountjoy, E.; Carmona, M.; Peat, G.; Schmidt, E.M.; Hercules, A.; Fumis, L.; Miranda, A.; Carvalho-Silva, D.; Buniello, A.; et al. Open Targets Genetics: Systematic identification of trait-associated genes using large-scale genetics and functional genomics. Nucleic Acids Res. 2020, 49, D1311–D1320. [Google Scholar] [CrossRef] [PubMed]
- Hormozdiari, F.; van de Bunt, M.; Segrè, A.V.; Li, X.; Joo, J.W.J.; Bilow, M.; Sul, J.H.; Sankararaman, S.; Pasaniuc, B.; Eskin, E. Colocalization of GWAS and eQTL Signals Detects Target Genes. Am. J. Hum. Genet. 2016, 99, 1245–1260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Landt, S.G.; Marinov, G.K.; Kundaje, A.; Kheradpour, P.; Pauli, F.; Batzoglou, S.; Bernstein, B.E.; Bickel, P.; Brown, J.B.; Cayting, P. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012, 22, 1813–1831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jayaram, N.; Usvyat, D.; Martin, A.C.R. Evaluating tools for transcription factor binding site prediction. BMC Bioinform. 2016, 17, 547. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- GTEx Portal. Available online: https://www.gtexportal.org/home/datasets (accessed on 22 December 2021).
- Fornes, O.; Castro-Mondragon, J.A.; Khan, A.; Van Der Lee, R.; Zhang, X.; Richmond, P.A.; Modi, B.P.; Correard, S.; Gheorghe, M.; Baranašić, D.; et al. JASPAR 2020: Update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2020, 48, D87–D92. [Google Scholar] [CrossRef] [PubMed]
- Kulakovskiy, I.V.; Vorontsov, I.E.; Yevshin, I.S.; Sharipov, R.N.; Fedorova, A.D.; Rumynskiy, E.I.; Medvedeva, Y.A.; Magana-Mora, A.; Bajic, V.B.; Papatsenko, D.A.; et al. HOCOMOCO: Towards a complete collection of transcription factor binding models for human and mouse via large-scale ChIP-Seq analysis. Nucleic Acids Res. 2017, 46, D252–D259. [Google Scholar] [CrossRef] [PubMed]
- Sebastian, A.; Contreras-Moreira, B. footprintDB: A database of transcription factors with annotated cis elements and binding interfaces. Bioinformatics 2013, 30, 258–265. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Hu, S.; Blackshaw, S.; Zhu, H.; Qian, J. hPDI: A database of experimental human protein-DNA interactions. Bioinformatics 2009, 26, 287–289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, D.; Wang, Z.; Zhou, Y.; Liang, Q.; Sham, P.C.; Yao, H.; Li, M.J. vSampler: Fast and annotation-based matched variant sampling tool. Bioinformatics 2020, 37, 1915–1917. [Google Scholar] [CrossRef] [PubMed]
- Ernst, J.; Kellis, M. Chromatin-state discovery and genome annotation with ChromHMM. Nat. Protoc. 2017, 12, 2478–2492. [Google Scholar] [CrossRef] [PubMed]
- Annotation of the non-coding genome. Nature 2015. [CrossRef] [Green Version]
- Zhang, Q.; Liu, W.; Zhang, H.-M.; Xie, G.-Y.; Miao, Y.-R.; Xia, M.; Guo, A.-Y. hTFtarget: A Comprehensive Database for Regulations of Human Transcription Factors and Their Targets. Genom. Proteom. Bioinform. 2020, 18, 120–128. [Google Scholar] [CrossRef] [PubMed]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. g: Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piñero, J.; Ramírez-Anguita, J.M.; Saüch-Pitarch, J.; Ronzano, F.; Centeno, E.; Sanz, F.; Furlong, L.I. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 2019, 48, D845–D855. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Disgenet Plus. Available online: https://beta.disgenetplus.com/ (accessed on 21 December 2021).
- Shadrina, M.; Bondarenko, E.A.; Slominsky, P.A. Genetics Factors in Major Depression Disease. Front. Psychiatry 2018, 9, 334. [Google Scholar] [CrossRef] [Green Version]
- McIntosh, A.M.; Sullivan, P.F.; Lewis, C.M. Uncovering the Genetic Architecture of Major Depression. Neuron 2019, 102, 91–103. [Google Scholar] [CrossRef]
- Zhang, K.; Lin, W.; Zhang, J.; Zhao, Y.; Wang, X.; Zhao, M. Effect of Toll-like receptor 4 on depressive-like behaviors induced by chronic social defeat stress. Brain Behav. 2020, 10, e01525. [Google Scholar] [CrossRef]
- Keyes, K.; Agnew-Blais, J.; Roberts, A.L.; Hamilton, A.; De Vivo, I.; Ranu, H.; Koenen, K. The role of allelic variation in estrogen receptor genes and major depression in the Nurses Health Study. Soc. Psychiatry 2015, 50, 1893–1904. [Google Scholar] [CrossRef] [Green Version]
- Mossakowska-Wójcik, J.; Orzechowska, A.; Talarowska, M.; Szemraj, J.; Gałecki, P. The importance of TCF4 gene in the etiology of recurrent depressive disorders. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2018, 80, 304–308. [Google Scholar] [CrossRef] [PubMed]
- Berrio, A.T.; Lopez, J.P.; Bagot, R.C.; Nouel, D.; Bo, G.D.; Cuesta, S.; Zhu, L.; Manitt, C.; Eng, C.; Cooper, H.M.; et al. DCC Confers Susceptibility to Depression-like Behaviors in Humans and Mice and Is Regulated by miR-218. Biol. Psychiatry 2016, 81, 306–315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hyde, C.L.; Nagle, M.W.; Tian, C.; Chen, X.; Paciga, S.A.; Wendland, J.R.; Tung, J.Y.; Hinds, D.A.; Perlis, R.H.; Winslow, A.R. Identification of 15 genetic loci associated with risk of major depression in individuals of European descent. Nat. Genet. 2016, 48, 1031–1036. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Tang, W.; Luo, Z.; Li, Y.; Shu, Y.; Yue, Z.; Xiao, B.; Feng, L. DISC1 Regulates the Proliferation and Migration of Mouse Neural Stem/Progenitor Cells through Pax5, Sox2, Dll1 and Neurog2. Front. Cell. Neurosci. 2017, 11, 261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rudzinskas, S.; Hoffman, J.F.; Martinez, P.; Rubinow, D.R.; Schmidt, P.J.; Goldman, D. In vitro model of perimenopausal depression implicates steroid metabolic and proinflammatory genes. Mol. Psychiatry 2020, 26, 3266–3276. [Google Scholar] [CrossRef]
- Qin, Z.; Ren, F.; Xu, X.; Ren, Y.; Li, H.; Wang, Y.; Zhai, Y.; Chang, Z. ZNF536, a Novel Zinc Finger Protein Specifically Expressed in the Brain, Negatively Regulates Neuron Differentiation by Repressing Retinoic Acid-Induced Gene Transcription. Mol. Cell. Biol. 2009, 29, 3633–3643. [Google Scholar] [CrossRef] [Green Version]
- Laifenfeld, D.; Klein, E.; Ben-Shachar, D. Norepinephrine alters the expression of genes involved in neuronal sprouting and differentiation: Relevance for major depression and antidepressant mechanisms. J. Neurochem. 2002, 83, 1054–1064. [Google Scholar] [CrossRef]
- Lanshakov, D.A.; Sukhareva, E.V.; Bulygina, V.V.; Bannova, A.V.; Shaburova, E.V.; Kalinina, T.S. Single neonatal dexamethasone administration has long-lasting outcome on depressive-like behaviour, Bdnf, Nt-3, p75ngfr and sorting receptors (SorCS1-3) stress reactive expression. Sci. Rep. 2021, 11, 8092. [Google Scholar] [CrossRef]
- Sanna, M.D.; Quattrone, A.; Galeotti, N. Antidepressant-like actions by silencing of neuronal ELAV-like RNA-binding proteins HuB and HuC in a model of depression in male mice. Neuropharmacology 2018, 135, 444–454. [Google Scholar] [CrossRef]
- Beilina, A.; Rudenko, I.N.; Kaganovich, A.; Civiero, L.; Chau, H.; Kalia, S.K.; Kalia, L.V.; Lobbestael, E.; Chia, R.; Ndukwe, K.; et al. Unbiased screen for interactors of leucine-rich repeat kinase 2 supports a common pathway for sporadic and familial Parkinson disease. Proc. Natl. Acad. Sci. USA 2014, 111, 2626–2631. [Google Scholar] [CrossRef] [Green Version]
- Dzhala, V.I.; Talos, D.M.; Sdrulla, D.A.; Brumback, A.; Mathews, G.C.; Benke, T.; Delpire, E.; Jensen, F.E.; Staley, K.J. NKCC1 transporter facilitates seizures in the developing brain. Nat. Med. 2005, 11, 1205–1213. [Google Scholar] [CrossRef] [PubMed]
- Bujalka, H.; Koenning, M.; Jackson, S.; Perreau, V.M.; Pope, B.; Hay, C.M.; Mitew, S.; Hill, A.F.; Lu, Q.R.; Wegner, M.; et al. MYRF Is a Membrane-Associated Transcription Factor That Autoproteolytically Cleaves to Directly Activate Myelin Genes. PLoS Biol. 2013, 11, e1001625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cross-Disorder Group of the Psychiatric Genomics Consortium. Genomic Relationships, Novel Loci, and Pleiotropic Mechanisms across Eight Psychiatric Disorders. Cell 2019, 179, 1469–1482.e11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, X.; Glessner, J.T.; Li, J.; Qi, X.; Hou, X.; Zhu, C.; Li, X.; March, M.E.; Yang, L.; Mentch, F.D.; et al. Integrative analysis of genome-wide association studies identifies novel loci associated with neuropsychiatric disorders. Transl. Psychiatry 2021, 11, 69. [Google Scholar] [CrossRef] [PubMed]
- Garcia, W.S.; Rocha-Acevedo, M.; Ramirez-Navarro, L.; Mbouamboua, Y.; Thieffry, D.; Thomas-Chollier, M.; Contreras-Moreira, B.; van Helden, J.; Medina-Rivera, A. RSAT variation-tools: An accessible and flexible framework to predict the impact of regulatory variants on transcription factor binding. Comput. Struct. Biotechnol. J. 2019, 17, 1415–1428. [Google Scholar] [CrossRef] [PubMed]
- Verheul, T.C.J.; Van Hijfte, L.; Perenthaler, E.; Barakat, T.S. The Why of YY1: Mechanisms of Transcriptional Regulation by Yin Yang 1. Front. Cell Dev. Biol. 2020, 8, 592164. [Google Scholar] [CrossRef]
- Byts, N.; Sharma, S.; Laurila, J.; Paudel, P.; Miinalainen, I.; Ronkainen, V.-P.; Hinttala, R.; Törnquist, K.; Koivunen, P.; Myllyharju, J. Transmembrane Prolyl 4-Hydroxylase is a Novel Regulator of Calcium Signaling in Astrocytes. ENeuro 2020, 8, 1–23. [Google Scholar] [CrossRef]
- Leinonen, H.; Koivisto, H.; Lipponen, H.-R.; Matilainen, A.; Salo, A.M.; Dimova, E.Y.; Hämäläinen, E.; Stavén, S.; Miettinen, P.; Myllyharju, J.; et al. Null mutation in P4h-tm leads to decreased fear and anxiety and increased social behavior in mice. Neuropharmacology 2019, 153, 63–72. [Google Scholar] [CrossRef]
- Bhalala, O.G.; Nath, A.P.; Inouye, M.; Sibley, C.R. UK Brain Expression Consortium Identification of expression quantitative trait loci associated with schizophrenia and affective disorders in normal brain tissue. PLoS Genet. 2018, 14, e1007607. [Google Scholar] [CrossRef]
- Li, S.; Li, X.; Liu, J.; Huo, Y.; Li, L.; Wang, J.; Luo, X.-J. Functional variants fine-mapping and gene function characterization provide insights into the role of ZNF323 in schizophrenia pathogenesis. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2021, 186, 28–39. [Google Scholar] [CrossRef]
- Roksana, Z. Transcription Factors in Schizophrenia: A Current View of Genetic Aspects. Sci. J. Genet. Gene Ther. 2016, 2, 17–21. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Su, X.; Liu, J.; Li, H.; Li, M.; Li, W.; Luo, X.-J. Transcriptome-wide association study identifies new susceptibility genes and pathways for depression. Transl. Psychiatry 2021, 11, 306. [Google Scholar] [CrossRef] [PubMed]
- Zhong, J.; Li, S.; Zeng, W.; Li, X.; Gu, C.; Liu, J.; Luo, X.-J. Integration of GWAS and brain eQTL identifies FLOT1 as a risk gene for major depressive disorder. Neuropsychopharmacology 2019, 44, 1542–1551. [Google Scholar] [CrossRef]
- Santos-Terra, J.; Deckmann, I.; Fontes-Dutra, M.; Schwingel, G.B.; Bambini-Junior, V.; Gottfried, C. Transcription factors in neurodevelopmental and associated psychiatric disorders: A potential convergence for genetic and environmental risk factors. Int. J. Dev. Neurosci. 2021, 81, 545–578. [Google Scholar] [CrossRef] [PubMed]
- Burt, C.; Munafò, M. Has GWAS lost its status as a paragon of open science? PLoS Biol. 2021, 19, e3001242. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.; Yao, X.; Shen, L. Integrative analysis of summary data from GWAS and eQTL studies implicates genes differentially expressed in Alzheimer’s disease. BMC Genom. 2022, 23, 414. [Google Scholar] [CrossRef] [PubMed]
- Brooks-Warburton, J.; Modos, D.; Sudhakar, P.; Madgwick, M.; Thomas, J.P.; Bohar, B.; Fazekas, D.; Zoufir, A.; Kapuy, O.; Szalay-Beko, M.; et al. A systems genomics approach to uncover patient-specific pathogenic pathways and proteins in ulcerative colitis. Nat. Commun. 2022, 13, 2299. [Google Scholar] [CrossRef]
- O’Brien, H.E.; Hannon, E.; Hill, M.J.; Toste, C.C.; Robertson, M.J.; Morgan, J.E.; McLaughlin, G.; Lewis, C.M.; Schalkwyk, L.C.; Hall, L.S.; et al. Expression quantitative trait loci in the developing human brain and their enrichment in neuropsychiatric disorders. Genome Biol. 2018, 19, 194. [Google Scholar] [CrossRef]
- Hare, B.D.; Duman, R.S. Prefrontal cortex circuits in depression and anxiety: Contribution of discrete neuronal populations and target regions. Mol. Psychiatry 2020, 25, 2742–2758. [Google Scholar] [CrossRef]
- Amare, A.T.; Vaez, A.; Hsu, Y.-H.; Direk, N.; Kamali, Z.; Howard, D.; McIntosh, A.; Tiemeier, H.; Bültmann, U.; Snieder, H.; et al. Bivariate genome-wide association analyses of the broad depression phenotype combined with major depressive disorder, bipolar disorder or schizophrenia reveal eight novel genetic loci for depression. Mol. Psychiatry 2019, 25, 1420–1429. [Google Scholar] [CrossRef]
- Pozzi, D.; Rasile, M.; Corradini, I.; Matteoli, M. Environmental regulation of the chloride transporter KCC2: Switching inflammation off to switch the GABA on? Transl. Psychiatry 2020, 10, 349. [Google Scholar] [CrossRef] [PubMed]
- Saponaro, F.; Sestito, S.; Runfola, M.; Rapposelli, S.; Chiellini, G. Selective Thyroid Hormone Receptor-Beta (TRβ) Agonists: New Perspectives for the Treatment of Metabolic and Neurodegenerative Disorders. Front. Med. 2020, 7, 331. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Zhu, Z.; Ransom, B.R.; Tong, X. Oligodendrocyte lineage cells and depression. Mol. Psychiatry 2020, 26, 103–117. [Google Scholar] [CrossRef] [PubMed]
- Lambert, S.A.; Jolma, A.; Campitelli, L.F.; Das, P.K.; Yin, Y.; Albu, M.; Chen, X.; Taipale, J.; Hughes, T.R.; Weirauch, M.T. The Human Transcription Factors. Cell 2018, 172, 650–665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andersen, M.C.; Engström, P.G.; Lithwick, S.; Arenillas, D.; Eriksson, P.; Lenhard, B.; Wasserman, W.W.; Odeberg, J. In Silico Detection of Sequence Variations Modifying Transcriptional Regulation. PLoS Comput. Biol. 2008, 4, e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dennis, D.J.; Han, S.; Schuurmans, C. bHLH transcription factors in neural development, disease, and reprogramming. Brain Res. 2019, 1705, 48–65. [Google Scholar] [CrossRef]
- Rada-Iglesias, A.; Ameur, A.; Kapranov, P.; Enroth, S.; Komorowski, J.; Gingeras, T.R.; Wadelius, C. Whole-genome maps of USF1 and USF2 binding and histone H3 acetylation reveal new aspects of promoter structure and candidate genes for common human disorders. Genome Res. 2008, 18, 380–392. [Google Scholar] [CrossRef] [Green Version]
- Sertbaş, M.; Ülgen, K.; Çakır, T. Systematic analysis of transcription-level effects of neurodegenerative diseases on human brain metabolism by a newly reconstructed brain-specific metabolic network. FEBS Open Bio 2014, 4, 542–553. [Google Scholar] [CrossRef] [Green Version]
- Grubert, F.; Srivas, R.; Spacek, D.V.; Kasowski, M.; Ruiz-Velasco, M.; Sinnott-Armstrong, N.; Greenside, P.; Narasimha, A.; Liu, Q.; Geller, B.; et al. Landscape of cohesin-mediated chromatin loops in the human genome. Nature 2020, 583, 737–743. [Google Scholar] [CrossRef]
- Brodie, A.; Azaria, J.R.; Ofran, Y. How far from the SNP may the causative genes be? Nucleic Acids Res. 2016, 44, 6046–6054. [Google Scholar] [CrossRef]
- Shi, Y.; Wang, Q.; Song, R.; Kong, Y.; Zhang, Z. Non-coding RNAs in depression: Promising diagnostic and therapeutic biomarkers. EBioMedicine 2021, 71, 103569. [Google Scholar] [CrossRef] [PubMed]
- Żurawek, D.; Turecki, G. The miRNome of Depression. Int. J. Mol. Sci. 2021, 22, 11312. [Google Scholar] [CrossRef] [PubMed]
- Bian, Z.; Li, H.; Liu, Y.; Cao, Y.; Kang, Y.; Yu, Y.; Zhang, F.; Li, C.; Kang, Y.; Wang, F. The Association Between Hypoxia Improvement and Electroconvulsive Therapy for Major Depressive Disorder. Neuropsychiatr. Dis. Treat. 2021, 17, 2987–2994. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Zhao, M.; Cheng, X.; Zhao, T.; Feng, Z.; Zhao, Y.; Fan, M.; Zhu, L. FG-4592 Improves Depressive-Like Behaviors through HIF-1-Mediated Neurogenesis and Synapse Plasticity in Rats. Neurotherapeutics 2019, 17, 664–675. [Google Scholar] [CrossRef] [PubMed]
- Ding, F.-S.; Cheng, X.; Zhao, T.; Zhao, Y.; Zhang, G.-B.; Wu, H.-T.; Zhu, L.-L.; Wu, K.-W. Intermittent hypoxic preconditioning relieves fear and anxiety behavior in post-traumatic stress model mice. Sheng Li Xue Bao 2019, 71, 537–546. [Google Scholar]
- Shibata, T.; Yamagata, H.; Uchida, S.; Otsuki, K.; Hobara, T.; Higuchi, F.; Abe, N.; Watanabe, Y. The alteration of hypoxia inducible factor-1 (HIF-1) and its target genes in mood disorder patients. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2013, 43, 222–229. [Google Scholar] [CrossRef] [PubMed]
- Kang, I.; Kondo, D.; Kim, J.; Lyoo, I.K.; Yurgelun-Todd, D.; Hwang, J.; Renshaw, P.F. Elevating the level of hypoxia inducible factor may be a new potential target for the treatment of depression. Med. Hypotheses 2020, 146, 110398. [Google Scholar] [CrossRef]
- Szczepocka, E.; Wysokiński, A. Red Blood Cells Parameters in Patients with Acute Schizophrenia, Unipolar Depression and Bipolar Disorder. Psychiatr. Danub. 2018, 30, 323–330. [Google Scholar] [CrossRef]





| GV | eGene | Tissue | PICS Probability GWAS | PICS Probability eQTL | Colocalization Probability |
|---|---|---|---|---|---|
| rs10149470 | BAG5 | Artery Tibial | 0.9657 | 0.633 | 0.6112881 |
| rs10149470 | RP11-894P9.2 | Colon Sigmoid | 0.9657 | 0.633 | 0.6112881 |
| rs10149470 | RP11-894P9.2 | Esophagus Gastroesophageal Junction | 0.9657 | 0.633 | 0.6112881 |
| rs10149470 | RP11-894P9.2 | Esophagus Muscularis | 0.9657 | 0.584 | 0.5639688 |
| rs10149470 | RP11-894P9.2 | Artery Aorta | 0.9657 | 0.499 | 0.4818843 |
| rs10149470 | RP11-894P9.2 | Breast Mammary Tissue | 0.9657 | 0.4494 | 0.43398558 |
| rs12624433 | SLC12A5 | Brain Putamen Basal Ganglia | 0.7355 | 0.303 | 0.2228565 |
| rs10149470 | RP11-894P9.2 | Stomach | 0.9657 | 0.1782 | 0.17208774 |
| rs10149470 | RP11-894P9.2 | Adipose Subcutaneous | 0.9657 | 0.1621 | 0.15653997 |
| rs10149470 | RP11-894P9.2 | Colon Transverse | 0.9657 | 0.1419 | 0.13703283 |
| rs10149470 | RP11-894P9.2 | Adipose Visceral Omentum | 0.9657 | 0.1412 | 0.13635684 |
| rs198457 | MYRF | Thyroid | 0.9627 | 0.1258 | 0.12110766 |
| rs10149470 | RP11-894P9.2 | Heart Left Ventricle | 0.9657 | 0.1225 | 0.11829825 |
| rs301799 | RP5-1115A15.1 | Whole Blood | 0.6946 | 0.1542 | 0.10710732 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pérez-Granado, J.; Piñero, J.; Medina-Rivera, A.; Furlong, L.I. Functional Genomics Analysis to Disentangle the Role of Genetic Variants in Major Depression. Genes 2022, 13, 1259. https://0-doi-org.brum.beds.ac.uk/10.3390/genes13071259
Pérez-Granado J, Piñero J, Medina-Rivera A, Furlong LI. Functional Genomics Analysis to Disentangle the Role of Genetic Variants in Major Depression. Genes. 2022; 13(7):1259. https://0-doi-org.brum.beds.ac.uk/10.3390/genes13071259
Chicago/Turabian StylePérez-Granado, Judith, Janet Piñero, Alejandra Medina-Rivera, and Laura I. Furlong. 2022. "Functional Genomics Analysis to Disentangle the Role of Genetic Variants in Major Depression" Genes 13, no. 7: 1259. https://0-doi-org.brum.beds.ac.uk/10.3390/genes13071259