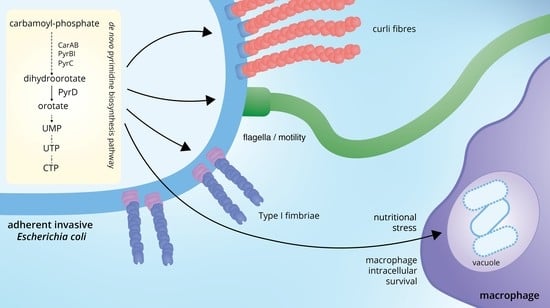
Inactivation of the Pyrimidine Biosynthesis pyrD Gene Negatively Affects Biofilm Formation and Virulence Determinants in the Crohn’s Disease-Associated Adherent Invasive Escherichia coli LF82 Strain
Abstract
:1. Introduction
2. Materials and Methods
2.1. Transposon Mutagenesis, Mutant Identification, and Screening on Acidic and Nutrient-Poor Medium
2.2. Adhesion Factor Detection, Biofilm Quantification, Motility Assay, MIC Determination and LPS Integrity Evaluation
2.3. Gene Expression Determination by Quantitative Real-Time PCR
2.4. Computational Models for Vidofludimus Binding to Dihydroorotate Dehydrogenase (DHOD) from Different Organisms
2.5. Statistical Analysis
3. Results
3.1. Mutant Selection in an Acidic and Nutrient Stress Medium Mimicking the Macrophage Vacuole Environment
3.2. The LF82pyrD::Tn5 Mutant Is Impaired in Biofilm Formation and Adhesion Factors’ Production and Displays a Slightly Reduced Flagellar Motility
3.3. The pyrD::Tn5 Mutation Results in Transcription Downregulation of Genes Encoding Curli Fibers and Type 1 Fimbriae
3.4. Dihydroorotate Dehydrogenase (DHOD) as Potential Drug Target
3.5. In Silico Analysis of Vidofludimus/DHOD Interaction
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- de Souza, H.S.P.; Fiocchi, C. Immunopathogenesis of IBD: Current State of the Art. Nat. Rev. Gastroenterol. 2016, 13, 13–27. [Google Scholar] [CrossRef] [PubMed]
- Duerr, R.H.; Taylor, K.D.; Brant, S.R.; Rioux, J.D.; Silverberg, M.S.; Daly, M.J.; Steinhart, A.H.; Abraham, C.; Regueiro, M.; Griffiths, A.; et al. A Genome-Wide Association Study Identifies IL23R as an Inflammatory Bowel Disease Gene. Science 2006, 314, 1461–1463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Villani, A.-C.; Lemire, M.; Fortin, G.; Louis, E.; Silverberg, M.S.; Collette, C.; Baba, N.; Libioulle, C.; Belaiche, J.; Bitton, A.; et al. Common Variants in the NLRP3 Region Contribute to Crohn’s Disease Susceptibility. Nat. Genet. 2009, 41, 71–76. [Google Scholar] [CrossRef] [PubMed]
- Ng, S.C.; Bernstein, C.N.; Vatn, M.H.; Lakatos, P.L.; Loftus, E.V.; Tysk, C.; O’Morain, C.; Moum, B.; Colombel, J.-F.; on behalf of the Epidemiology and Natural History Task Force of the International Organization of Inflammatory Bowel Disease (IOIBD). Geographical Variability and Environmental Risk Factors in Inflammatory Bowel Disease. Gut 2013, 62, 630. [Google Scholar] [CrossRef]
- Wang, K.; Wu, L.; Dou, C.; Guan, X.; Wu, H.; Liu, H. Research Advance in Intestinal Mucosal Barrier and Pathogenesis of Crohn’s Disease. Gastroenterol. Res. Pract. 2016, 2016, 9686238. [Google Scholar] [CrossRef] [Green Version]
- Geremia, A.; Biancheri, P.; Allan, P.; Corazza, G.R.; Sabatino, A.D. Innate and Adaptive Immunity in Inflammatory Bowel Disease. Autoimmun. Rev. 2014, 13, 3–10. [Google Scholar] [CrossRef]
- Ahmed, I.; Roy, B.C.; Khan, S.A.; Septer, S.; Umar, S. Microbiome, Metabolome and Inflammatory Bowel Disease. Microorganisms 2016, 4, 20. [Google Scholar] [CrossRef] [Green Version]
- Nishida, A.; Inoue, R.; Inatomi, O.; Bamba, S.; Naito, Y.; Andoh, A. Gut Microbiota in the Pathogenesis of Inflammatory Bowel Disease. Clin. J. Gastroenterol. 2018, 11, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Walters, W.A.; Xu, Z.; Knight, R. Meta-analyses of Human Gut Microbes Associated with Obesity and IBD. FEBS Lett. 2014, 588, 4223–4233. [Google Scholar] [CrossRef] [Green Version]
- Gevers, D.; Kugathasan, S.; Knights, D.; Kostic, A.D.; Knight, R.; Xavier, R.J. A Microbiome Foundation for the Study of Crohn’s Disease. Cell Host Microbe 2017, 21, 301–304. [Google Scholar] [CrossRef] [Green Version]
- Mentella, M.C.; Scaldaferri, F.; Pizzoferrato, M.; Gasbarrini, A.; Miggiano, G.A.D. Nutrition, IBD and Gut Microbiota: A Review. Nutrients 2020, 12, 944. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Darfeuille-Michaud, A.; Boudeau, J.; Bulois, P.; Neut, C.; Glasser, A.-L.; Barnich, N.; Bringer, M.-A.; Swidsinski, A.; Beaugerie, L.; Colombel, J.-F. High Prevalence of Adherent-Invasive Escherichia coli Associated with Ileal Mucosa in Crohn’s Disease. Gastroenterology 2004, 127, 412–421. [Google Scholar] [CrossRef] [PubMed]
- Schippa, S.; Conte, M.P.; Borrelli, O.; Iebba, V.; Aleandri, M.; Seganti, L.; Longhi, C.; Chiarini, F.; Osborn, J.; Cucchiara, S. Dominant Genotypes in Mucosa-Associated Escherichia coli Strains from Pediatric Patients with Inflammatory Bowel Disease. Inflamm. Bowel Dis. 2009, 15, 661–672. [Google Scholar] [CrossRef] [PubMed]
- Thomazini, C.M.; Samegima, D.A.G.; Rodrigues, M.A.M.; Victoria, C.R.; Rodrigues, J. High Prevalence of Aggregative Adherent Escherichia coli Strains in the Mucosa-Associated Microbiota of Patients with Inflammatory Bowel Diseases. Int. J. Med. Microbiol. 2011, 301, 475–479. [Google Scholar] [CrossRef] [PubMed]
- Glasser, A.-L.; Boudeau, J.; Barnich, N.; Perruchot, M.-H.; Colombel, J.-F.; Darfeuille-Michaud, A. Adherent Invasive Escherichia coli Strains from Patients with Crohn’s Disease Survive and Replicate within Macrophages without Inducing Host Cell Death. Infect. Immun. 2001, 69, 5529–5537. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eaves-Pyles, T.; Allen, C.A.; Taormina, J.; Swidsinski, A.; Tutt, C.B.; Jezek, G.E.; Islas-Islas, M.; Torres, A.G. Escherichia coli Isolated from a Crohn’s Disease Patient Adheres, Invades, and Induces Inflammatory Responses in Polarized Intestinal Epithelial Cells. Int. J. Med. Microbiol. 2008, 298, 397–409. [Google Scholar] [CrossRef] [PubMed]
- Rolhion, N.; Darfeuille-Michaud, A. Adherent-Invasive Escherichia coli in Inflammatory Bowel Disease. Inflamm. Bowel Dis. 2007, 13, 1277–1283. [Google Scholar] [CrossRef]
- Boudeau, J.; Barnich, N.; Darfeuille-Michaud, A. Type 1 Pili-mediated Adherence of Escherichia coli Strain LF82 Isolated from Crohn’s Disease Is Involved in Bacterial Invasion of Intestinal Epithelial Cells. Mol. Microbiol. 2001, 39, 1272–1284. [Google Scholar] [CrossRef]
- Barnich, N.; Boudeau, J.; Claret, L.; Darfeuille-Michaud, A. Regulatory and Functional Co-operation of Flagella and Type 1 Pili in Adhesive and Invasive Abilities of AIEC Strain LF82 Isolated from a Patient with Crohn’s Disease. Mol. Microbiol. 2003, 48, 781–794. [Google Scholar] [CrossRef]
- Cieza, R.J.; Hu, J.; Ross, B.N.; Sbrana, E.; Torres, A.G. The IbeA Invasin of Adherent-Invasive Escherichia coli Mediates Interaction with Intestinal Epithelia and Macrophages. Infect. Immun. 2015, 83, 1904–1918. [Google Scholar] [CrossRef] [Green Version]
- Chassaing, B.; Darfeuille-Michaud, A. The σE Pathway Is Involved in Biofilm Formation by Crohn’s Disease-Associated Adherent-Invasive Escherichia coli. J. Bacteriol. 2013, 195, 76–84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fanelli, G.; Pasqua, M.; Colonna, B.; Prosseda, G.; Grossi, M. Expression Profile of Multidrug Resistance Efflux Pumps During Intracellular Life of Adherent-Invasive Escherichia coli Strain LF82. Front. Microbiol. 2020, 11, 1935. [Google Scholar] [CrossRef] [PubMed]
- D’Haens, G.R.; Vermeire, S.; Assche, G.V.; Noman, M.; Aerden, I.; Olmen, G.V.; Rutgeerts, P. Therapy of Metronidazole With Azathioprine to Prevent Postoperative Recurrence of Crohn’s Disease: A Controlled Randomized Trial. Gastroenterology 2008, 135, 1123–1129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shin, S.J.; Collins, M.T. Thiopurine Drugs Azathioprine and 6-Mercaptopurine Inhibit Mycobacterium paratuberculosis Growth In Vitro. Antimicrob. Agents Chemother. 2008, 52, 418–426. [Google Scholar] [CrossRef] [Green Version]
- Migliore, F.; Macchi, R.; Landini, P.; Paroni, M. Phagocytosis and Epithelial Cell Invasion by Crohn’s Disease-Associated Adherent-Invasive Escherichia coli Are Inhibited by the Anti-Inflammatory Drug 6-Mercaptopurine. Front. Microbiol. 2018, 9, 964. [Google Scholar] [CrossRef]
- Nitzan, O.; Elias, M.; Peretz, A.; Saliba, W. Role of Antibiotics for Treatment of Inflammatory Bowel Disease. World J. Gastroenterol. 2016, 22, 1078. [Google Scholar] [CrossRef]
- Ganji-Arjenaki, M.; Rafieian-Kopaei, M. Probiotics Are a Good Choice in Remission of Inflammatory Bowel Diseases: A Meta Analysis and Systematic Review. J. Cell Physiol. 2018, 233, 2091–2103. [Google Scholar] [CrossRef]
- Ghouri, Y.A.; Richards, D.M.; Rahimi, E.F.; Krill, J.T.; Jelinek, K.A.; DuPont, A.W. Systematic Review of Randomized Controlled Trials of Probiotics, Prebiotics, and Synbiotics in Inflammatory Bowel Disease. Clin. Exp. Gastroenterol. 2014, 7, 473–487. [Google Scholar] [CrossRef] [Green Version]
- Butterworth, A.D.; Thomas, A.G.; Akobeng, A.K. Probiotics for Induction of Remission in Crohn’s Disease. Cochrane Database Syst. Rev. 2008, CD006634. [Google Scholar] [CrossRef]
- Leccese, G.; Bibi, A.; Mazza, S.; Facciotti, F.; Caprioli, F.; Landini, P.; Paroni, M. Probiotic Lactobacillus and Bifidobacterium Strains Counteract Adherent-Invasive Escherichia coli (AIEC) Virulence and Hamper IL-23/Th17 Axis in Ulcerative Colitis, but Not in Crohn’s Disease. Cells 2020, 9, 1824. [Google Scholar] [CrossRef]
- Durand, J.M.B.; Björk, G.R. Metabolic Control through Ornithine and Uracil of Epithelial Cell Invasion by Shigella flexneri. Microbiology 2009, 155, 2498–2508. [Google Scholar] [CrossRef] [Green Version]
- Ueda, A.; Attila, C.; Whiteley, M.; Wood, T.K. Uracil Influences Quorum Sensing and Biofilm Formation in Pseudomonas aeruginosa and Fluorouracil Is an Antagonist. Microb. Biotechnol. 2009, 2, 62–74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boudeau, J.; Glasser, A.-L.; Masseret, E.; Joly, B.; Darfeuille-Michaud, A. Invasive Ability of an Escherichia coli Strain Isolated from the Ileal Mucosa of a Patient with Crohn’s Disease. Infect. Immun. 1999, 67, 4499–4509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bringer, M.-A.; Rolhion, N.; Glasser, A.-L.; Darfeuille-Michaud, A. The Oxidoreductase DsbA Plays a Key Role in the Ability of the Crohn’s Disease-Associated Adherent-Invasive Escherichia coli Strain LF82 To Resist Macrophage Killing. J. Bacteriol. 2007, 189, 4860–4871. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thompson, A.P.; O’Neill, I.; Smith, E.J.; Catchpole, J.; Fagan, A.; Burgess, K.E.V.; Carmody, R.J.; Clarke, D.J. Glycolysis and Pyrimidine Biosynthesis Are Required for Replication of Adherent–Invasive Escherichia coli in Macrophages. Microbiology 2016, 162, 954–965. [Google Scholar] [CrossRef] [PubMed]
- Garavaglia, M.; Rossi, E.; Landini, P. The Pyrimidine Nucleotide Biosynthetic Pathway Modulates Production of Biofilm Determinants in Escherichia coli. PLoS ONE 2012, 7, e31252. [Google Scholar] [CrossRef] [Green Version]
- Genevaux, P.; Bauda, P.; DuBow, M.S.; Oudega, B. Identification of Tn 10 Insertions in the RfaG, RfaP, and GalU Genes Involved in Lipopolysaccharide Core Biosynthesis That Affect Escherichia coli Adhesion. Arch. Microbiol. 1999, 172, 1–8. [Google Scholar] [CrossRef]
- Møller, A.K.; Leatham, M.P.; Conway, T.; Nuijten, P.J.M.; de Haan, L.A.; Krogfelt, K.A.; Cohen, P.S. An Escherichia coli MG1655 Lipopolysaccharide Deep-Rough Core Mutant Grows and Survives in Mouse Cecal Mucus but Fails To Colonize the Mouse Large Intestine. Infect. Immun. 2003, 71, 2142–2152. [Google Scholar] [CrossRef] [Green Version]
- Dartigalongue, C.; Missiakas, D.; Raina, S. Characterization of the Escherichia coli σE Regulon. J. Biol. Chem. 2001, 276, 20866–20875. [Google Scholar] [CrossRef] [Green Version]
- Tam, C.; Missiakas, D. Changes in Lipopolysaccharide Structure Induce the σE-dependent Response of Escherichia coli. Mol. Microbiol. 2005, 55, 1403–1412. [Google Scholar] [CrossRef]
- Römling, U.; Rohde, M.; Olsén, A.; Normark, S.; Reinköster, J. AgfD, the Checkpoint of Multicellular and Aggregative Behaviour in Salmonella typhimurium Regulates at Least Two Independent Pathways. Mol. Microbiol. 2000, 36, 10–23. [Google Scholar] [CrossRef] [PubMed]
- Zogaj, X.; Nimtz, M.; Rohde, M.; Bokranz, W.; Römling, U. The Multicellular Morphotypes of Salmonella typhimurium and Escherichia coli Produce Cellulose as the Second Component of the Extracellular Matrix. Mol. Microbiol. 2001, 39, 1452–1463. [Google Scholar] [CrossRef] [PubMed]
- Herrlinger, K.R.; Diculescu, M.; Fellermann, K.; Hartmann, H.; Howaldt, S.; Nikolov, R.; Petrov, A.; Reindl, W.; Otte, J.M.; Stoynov, S.; et al. Efficacy, Safety and Tolerability of Vidofludimus in Patients with Inflammatory Bowel Disease: The ENTRANCE Study. J. Crohn’s Colitis 2013, 7, 636–643. [Google Scholar] [CrossRef] [Green Version]
- Ellis, M.J.; Tsai, C.N.; Johnson, J.W.; French, S.; Elhenawy, W.; Porwollik, S.; Andrews-Polymenis, H.; McClelland, M.; Magolan, J.; Coombes, B.K.; et al. A Macrophage-Based Screen Identifies Antibacterial Compounds Selective for Intracellular Salmonella typhimurium. Nat. Commun. 2019, 10, 197. [Google Scholar] [CrossRef] [Green Version]
- Bange, F.C.; Brown, A.M.; Jacobs, W.R. Leucine Auxotrophy Restricts Growth of Mycobacterium bovis BCG in Macrophages. Infect. Immun. 1996, 64, 1794–1799. [Google Scholar] [CrossRef] [Green Version]
- Pilatz, S.; Breitbach, K.; Hein, N.; Fehlhaber, B.; Schulze, J.; Brenneke, B.; Eberl, L.; Steinmetz, I. Identification of Burkholderia pseudomallei Genes Required for the Intracellular Life Cycle and In Vivo Virulence. Infect. Immun. 2006, 74, 3576–3586. [Google Scholar] [CrossRef] [Green Version]
- Smith, D.A.; Parish, T.; Stoker, N.G.; Bancroft, G.J. Characterization of Auxotrophic Mutants of Mycobacterium tuberculosis and Their Potential as Vaccine Candidates. Infect. Immun. 2001, 69, 1142–1150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beaumont, H.J.E.; Gallie, J.; Kost, C.; Ferguson, G.C.; Rainey, P.B. Experimental Evolution of Bet Hedging. Nature 2009, 462, 90. [Google Scholar] [CrossRef] [Green Version]
- Tükel, Ç.; Nishimori, J.H.; Wilson, R.P.; Winter, M.G.; Keestra, A.M.; Putten, J.P.M.V.; Bäumler, A.J. Toll-like Receptors 1 and 2 Cooperatively Mediate Immune Responses to Curli, a Common Amyloid from Enterobacterial Biofilms. Cell Microbiol. 2010, 12, 1495–1505. [Google Scholar] [CrossRef] [Green Version]
- Rapsinski, G.J.; Wynosky-Dolfi, M.A.; Oppong, G.O.; Tursi, S.A.; Wilson, R.P.; Brodsky, I.E.; Tükel, Ç. Toll-like Receptor 2 and NLRP3 Cooperate to Recognize a Functional Bacterial Amyloid, Curli. Infect. Immun. 2014, 83, 693–701. [Google Scholar] [CrossRef] [Green Version]
- Lauritsen, I.; Frendorf, P.O.; Capucci, S.; Heyde, S.A.H.; Blomquist, S.D.; Wendel, S.; Fischer, E.C.; Sekowska, A.; Danchin, A.; Nørholm, M.H.H. Temporal Evolution of Master Regulator Crp Identifies Pyrimidines as Catabolite Modulator Factors. Nat. Commun. 2021, 12, 5880. [Google Scholar] [CrossRef] [PubMed]
- Rossi, E.; Motta, S.; Aliverti, A.; Cossu, F.; Gourlay, L.; Mauri, P.; Landini, P. Cellulose Production Is Coupled to Sensing of the Pyrimidine Biosynthetic Pathway via c-di-GMP Production by the DgcQ Protein of Escherichia coli. Environ. Microbiol. 2017, 19, 4551–4563. [Google Scholar] [CrossRef] [Green Version]
- Muehler, A.; Kohlhof, H.; Groeppel, M.; Vitt, D. Safety, Tolerability and Pharmacokinetics of Vidofludimus Calcium (IMU-838) After Single and Multiple Ascending Oral Doses in Healthy Male Subjects. Eur. J. Drug Metab. Pharmacokinet. 2020, 45, 557–573. [Google Scholar] [CrossRef] [PubMed]
- Antoniani, D.; Rossi, E.; Rinaldo, S.; Bocci, P.; Lolicato, M.; Paiardini, A.; Raffaelli, N.; Cutruzzolà, F.; Landini, P. The Immunosuppressive Drug Azathioprine Inhibits Biosynthesis of the Bacterial Signal Molecule Cyclic-Di-GMP by Interfering with Intracellular Nucleotide Pool Availability. Appl. Microbiol. Biot. 2013, 97, 7325–7336. [Google Scholar] [CrossRef] [PubMed]
- Leclercq, R.; Courvalin, P. Intrinsic and Unusual Resistance to Macrolide, Lincosamide, and Streptogramin Antibiotics in Bacteria. Antimicrob. Agents Chemother. 1991, 35, 1273–1276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nørager, S.; Jensen, K.F.; Björnberg, O.; Larsen, S. E. Coli Dihydroorotate Dehydrogenase Reveals Structural and Functional Distinctions between Different Classes of Dihydroorotate Dehydrogenases. Structure 2002, 10, 1211–1223. [Google Scholar] [CrossRef] [Green Version]
- Marcinkeviciene, J.; Rogers, M.J.; Kopcho, L.; Jiang, W.; Wang, K.; Murphy, D.J.; Lippy, J.; Link, S.; Chung, T.D.Y.; Hobbs, F.; et al. Selective Inhibition of Bacterial Dihydroorotate Dehydrogenases by Thiadiazolidinediones. Biochem. Pharmacol. 2000, 60, 339–342. [Google Scholar] [CrossRef]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rossi, E.; Leccese, G.; Baldelli, V.; Bibi, A.; Scalone, E.; Camilloni, C.; Paroni, M.; Landini, P. Inactivation of the Pyrimidine Biosynthesis pyrD Gene Negatively Affects Biofilm Formation and Virulence Determinants in the Crohn’s Disease-Associated Adherent Invasive Escherichia coli LF82 Strain. Microorganisms 2022, 10, 537. https://0-doi-org.brum.beds.ac.uk/10.3390/microorganisms10030537
Rossi E, Leccese G, Baldelli V, Bibi A, Scalone E, Camilloni C, Paroni M, Landini P. Inactivation of the Pyrimidine Biosynthesis pyrD Gene Negatively Affects Biofilm Formation and Virulence Determinants in the Crohn’s Disease-Associated Adherent Invasive Escherichia coli LF82 Strain. Microorganisms. 2022; 10(3):537. https://0-doi-org.brum.beds.ac.uk/10.3390/microorganisms10030537
Chicago/Turabian StyleRossi, Elio, Gabriella Leccese, Valerio Baldelli, Alessia Bibi, Emanuele Scalone, Carlo Camilloni, Moira Paroni, and Paolo Landini. 2022. "Inactivation of the Pyrimidine Biosynthesis pyrD Gene Negatively Affects Biofilm Formation and Virulence Determinants in the Crohn’s Disease-Associated Adherent Invasive Escherichia coli LF82 Strain" Microorganisms 10, no. 3: 537. https://0-doi-org.brum.beds.ac.uk/10.3390/microorganisms10030537