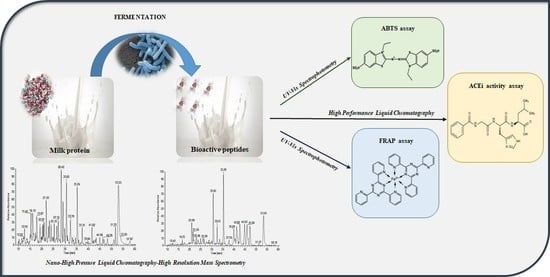
Chromatographic Characterization and In Vitro Bioactivity Evaluation of Lactobacillus helveticus Hydrolysates upon Fermentation of Different Substrates
Abstract
:Featured Application
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. HPLC-UV Conditions
2.3. Bacterial Strain Selection and Propagation
2.4. Evaluation of Different Substrates and Comparison of Two Extraction Protocols for Milk Fermented with L. helveticus
2.5. Determination of the ACE Inhibitory Activity
2.6. Determination of the Radical Scavenging Capacity by the ABTS Method
2.7. Determination of the Antioxidant Capacity by the Ferric Reducing Antioxidant Power (FRAP) Assay
2.8. Nano HPLC-HRMS Analysis
2.9. Statistical Analysis
3. Results and Discussion
3.1. Evaluation of the Fermentation Activity by RP-HPLC
3.2. Antioxidant and ACE-Inhibitory Activity of Peptide Hydrolysates
3.3. Profiling of SM Samples by Mass Spectrometry
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Beltrán-Barrientos, L.M.; Hernández-Mendoza, A.; Torres-Llanez, M.J.; González-Córdova, A.F.; Vallejo-Córdoba, B. Invited review: Fermented milk as antihypertensive functional food. J. Dairy Sci. 2015, 99, 4099–4110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prasanna, P.H.P.; Grandison, A.S.D.; Charalampopoulos, D. Bifidobacteria in milk products: An overview of physiological and biochemical properties, exopolysaccharide production, selection criteria of milk products and health benefits. Food Res. Int. 2014, 55, 247–262. [Google Scholar] [CrossRef]
- Chakrabarti, S.; Guha, S.; Majumder, K. Food-Derived Bioactive Peptides in Human Health: Challenges and Opportunities. Nutrients 2018, 10, 1738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montesano, D.; Gallo, M.; Blasi, F.; Cossignani, L. Biopeptides from vegetable proteins: New scientific evidences. Curr. Opin. Food Sci. 2020, 31, 31–37. [Google Scholar] [CrossRef]
- Rai, A.K.; Sanjukta, S.; Jeyaram, K. Production of angiotensin I converting enzyme inhibitory (ACE-I) peptides during milk fermentation and their role in reducing hypertension. Crit. Rev. Food Sci. Nutr. 2017, 57, 2789–2800. [Google Scholar] [CrossRef]
- Saito, T.; Nakamura, H.; Kitazawa, H.; Kawai, Y.; Itoh, T. Isolation and structural analysis of antihypertensive peptides that exist naturally in Gouda cheese. J. Dairy Sci. 2000, 83, 1434–1440. [Google Scholar] [CrossRef]
- Schanbacher, F.L.; Talhouk, R.S.; Murray, F.A.; Gherman, L.I.; Willet, L.B. Milk borne bioactive peptides. Int. Dairy J. 1998, 8, 393–403. [Google Scholar] [CrossRef]
- Hafeez, Z.; Cakir-Kiefer, C.; Roux, E.; Perrin, C.; Miclo, L.; Dary-Mourot, A. Strategies of producing bioactive peptides from milk proteins to functionalize fermented milk products. Food Res. Int. 2014, 63, 71–80. [Google Scholar] [CrossRef]
- Power, O.; Jakeman, P.; FitzGerald, R.J. Antioxidative peptides: Enzymatic production, in vitro and in vivo antioxidant activity and potential applications of milk-derived antioxidative peptides. Amino Acids 2013, 44, 797–820. [Google Scholar] [CrossRef]
- Rana, S.; Bajaj, R.; Mann, B. Characterization of antimicrobial and antioxidative peptides synthesized by L. rhamnosus C6 fermentation of milk. Int. J. Pept. Res. Ther. 2018, 24, 309–321. [Google Scholar] [CrossRef]
- Pihlanto, A.; Virtanen, T.; Korhonen, H. Angiotensin I converting enzyme (ACE) inhibitory activity and antihypertensive effect of fermented milk. Int. Dairy J. 2010, 20, 3–10. [Google Scholar] [CrossRef]
- Muguerza, B.; Ramos, M.; Sanchez, E.; Manso, M.A.; Miguel, M.; Aleixandre, A.; Delgado, M.A.; Recio, I. Antihypertensive activity of milk fermented by Enterococcus faecalis strains isolated from raw milk. Int. Dairy Res. 2006, 16, 61–69. [Google Scholar] [CrossRef]
- Wang, J.; Li, C.; Xue, J.; Yang, J.; Zhang, Q.; Zhang, H.; Chen, Y. Fermentation characteristics and angiotensin I-converting enzyme-inhibitory activity of Lactobacillus helveticus isolate H9 in cow milk, soy milk, and mare milk. J. Dairy Sci. 2015, 98, 3655–3664. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, M.U.; Pirzadeh, M.; Förster, C.Y.; Shityakov, S.; Shariati, M.A. Role of milk-derived antibacterial peptides in modern food biotechnology: Their synthesis, applications and future perspectives. Biomolecules 2018, 8, 110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davoodi, S.H.; Shahbazi, R.; Esmaeili, S.; Sohrabvandi, S.; Mortazavian, A.; Jazayeri, S.; Taslimi, A. Health-Related Aspects of Milk Proteins. Iran. J. Pharm. Res. 2016, 15, 573–591. [Google Scholar] [PubMed]
- Tagliazucchi, D.; Martini, S.; Solieri, L. Bioprospecting for bioactive peptide production by lactic acid bacteria isolated from fermented dairy food. Fermentation 2019, 5, 96. [Google Scholar] [CrossRef] [Green Version]
- Mohanty, D.; Jena, R.; Choudhury, P.K.; Pattnaik, R.; Mohapatra, S.; Saini, M.R. Milk derived antimicrobial bioactive peptides: A review. Int. J. Food Prop. 2016, 19, 837–846. [Google Scholar] [CrossRef]
- Cenci-Goga, B.T.; Karama, M.; Sechi, P.; Iulietto, M.F.; Grispoldi, L.; Selvaggini, R.; Ceccarelli, M.; Barbera, S. Fate of selected pathogens in spiked «SALAME NOSTRANO» produced without added nitrates following the application of NONIT™ technology. Meat Sci. 2018, 139, 247–254. [Google Scholar] [CrossRef]
- Cenci-Goga, B.T.; Karama, M.; Sechi, P.; Iulietto, M.F.; Novelli, S.; Selvaggini, R.; Barbera, S. Effect of a novel starter culture and specific ripening conditions on microbiological characteristics of nitrate-free dry-cured pork sausages. Ital. J. Anim. Sci. 2016, 15, 358–374. [Google Scholar] [CrossRef]
- Cenci-Goga, B.T.; Karama, M.; Sechi, P.; Iulietto, M.F.; Novelli, S.; Selvaggini, R.; Mattei, S. Growth inhibition of selected microorganisms by an association of dairy starter cultures and probiotics. Ital. J. Anim. Sci. 2015, 14, 246–250. [Google Scholar] [CrossRef] [Green Version]
- Cenci-Goga, B.T.; Rossitto, P.V.; Sechi, P.; Parmegiani, S.; Cambiotti, V.; Cullor, J.S. Effect of selected dairy starter cultures on microbiological, chemical and sensory characteristics of swine and venison (Damadama) nitrite-free dry-cured sausages. Meat Sci. 2012, 90, 599–606. [Google Scholar] [CrossRef] [PubMed]
- Clementi, F.; Cenci Goga, B.T.; Trabalza Marinucci, M.; Di Antonio, E. Use of selected starter cultures in the production of farm manufactured goat cheese from thermized milk. Ital. J. Food Sci. 1998, 10, 41–56. [Google Scholar]
- Urbani, E.; Blasi, F.; Simonetti, M.S.; Chiesi, C.; Cossignani, L. Investigation on secondary metabolite content and antioxidant activity of commercial saffron powder. Eur. Food Res. Technol. 2016, 242, 987–993. [Google Scholar] [CrossRef]
- Rocchetti, G.; Pagnossa, J.P.; Blasi, F.; Cossignani, L.; HilsdorfPiccoli, R.; Zengin, G.; Montesano, D.; Cocconcelli, P.S.; Lucini, L. Phenolic profiling and in vitro bioactivity of Moringa oleifera leaves as affected by different extraction solvents. Food Res. Int. 2020, 127, 108712. [Google Scholar] [CrossRef] [PubMed]
- Rocchetti, G.; Blasi, F.; Montesano, D.; Ghisoni, S.; Marcotullio, M.C.; Sabatini, S.; Cossignani, L.; Lucini, L. Impact of conventional/non-conventional extraction methods on the untargeted phenolic profile of Moringa oleifera leaves. Food Res. Int. 2019, 115, 319–327. [Google Scholar] [CrossRef] [PubMed]
- Degani, G.; Altomare, A.; Digiovanni, S.; Arosio, B.; Fritz, G.; Raucci, A.; Aldini, G.; Popolo, L. Prothrombin is a binding partner of the human receptor of advanced glycation end products. J. Biol. Chem. 2020, 295, 12498–12511. [Google Scholar] [CrossRef]
- Chen, G.W.; Tsai, J.S.; Pan, B.S. Purification of angiotensin I-converting enzyme inhibitory peptides and antihypertensive effect of milk produced by protease-facilitated lactic fermentation. Int. Dairy J. 2007, 17, 641–647. [Google Scholar] [CrossRef]
- Hernandez-Ledesma, B.; Amigo, L.; Ramos, M.; Recio, I. Application of high-performance liquid chromatography-tandem mass spectrometry to the identification of biologically active peptides produced by milk fermentation and simulated gastrointestinal digestion. J. Chromatogr. A 2004, 1049, 107–114. [Google Scholar] [CrossRef]
- Ianni, F.; Sardella, R.; Lisanti, A.; Gioiello, A.; Cenci Goga, B.T.; Lindner, W.; Natalini, B. Achiral-chiral two-dimensional chromatography of free amino acids in milk: A promising tool for detecting different levels of mastitis in cows. J. Pharm. Biomed. Anal. 2005, 116, 40–46. [Google Scholar] [CrossRef]
- Mant, C.T.; Hodges, R.S. HPLC of Biological Macromolecules; Gooding, K.M., Regnier, F.E., Eds.; Marcel Dekker: New York, NY, USA, 2002; pp. 433–511. [Google Scholar]
- Chen, Y.; Mehok, A.R.; Mant, C.T.; Hodges, R.S. Optimum concentration of trifluoroacetic acid for reversed-phase liquid chromatography of peptides revisited. J. Chromatogr. A 2004, 1043, 9–18. [Google Scholar] [CrossRef]
- Dolan, J.W.; Snyder, L.R.; Blanc, T.; Van Heukelem, L. Selectivity differences for C18 and C8 reversed-phase columns as a function of temperature and gradient steepness: I. Optimizing selectivity and resolution. J. Chromatogr. A 2000, 897, 37–50. [Google Scholar] [CrossRef]
- Hancock, W.S.; Chloupek, R.C.; Kirkland, J.J.; Snyder, L.R. Temperature as a variable in reversed-phase high-performance liquid chromatographic separations of peptide and protein samples: I. Optimizing the separation of a growth hormone tryptic digest. J. Chromatogr. A 1994, 686, 31–43. [Google Scholar] [CrossRef]
- Chloupek, R.C.; Hancock, W.S.; Marchylo, B.A.; Kirkland, J.J.; Boyes, B.E.; Snyder, L.R. Temperature as a variable in reversed-phase high-performance liquid chromatographic separations of peptide and protein samples. II. Selectivity effects observed in the separation of several peptide and protein mixtures. J. Chromatogr. A 1994, 686, 45–59. [Google Scholar] [CrossRef]
- Sardella, R.; Lisanti, A.; Marinozzi, M.; Ianni, F.; Natalini, B.; Blanch, G.P.; Ruiz del Castillo, M.L. Combined monodimensional chromatographic approaches to monitor the presence of D-amino acids in cheese. Food Control 2013, 34, 478–487. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Liu, W.; Xue, J.; Yang, J.; Chen, X.; Shao, Y.; Kwok, L.; Bilige, M.; Mang, L.; Zhang, H. Angiotensin-converting enzyme inhibitory activity of Lactobacillus helveticus strains from traditional fermented dairy foods and antihypertensive effect of fermented milk of strain H9. J. Dairy Sci. 2014, 97, 6680–6692. [Google Scholar] [CrossRef] [Green Version]
- Solieri, L.; Rutella, G.S.; Tagliazucchi, D. Impact of non-starter lactobacilli on release of peptides with angiotensin-converting enzyme inhibitory and antioxidant activities during bovine milk fermentation. Food Microbiol. 2015, 51, 108–116. [Google Scholar] [CrossRef] [Green Version]
- Pan, D.; Luo, Y.; Tanokura, M. Antihypertensive peptides from skimmed milk hydrolysate digested by cell-free extract of Lactobacillus helveticus JCM1004. Food Chem. 2005, 91, 123–129. [Google Scholar] [CrossRef]
- Li, B.; Chen, F.; Wang, X.; Ji, B.; Wu, Y. Isolation and identification of antioxidant peptides from porcine collagen hydrolysate by consecutive chromatography and electrospray ionization-mass spectrometry. Food Chem. 2007, 102, 1135–1143. [Google Scholar] [CrossRef]
- Sarmadi, B.H.; Ismail, A. Antioxidative peptides from food proteins: A review. Peptides 2010, 31, 1949–1956. [Google Scholar] [CrossRef]
- Chen, H.M.; Muramoto, K.; Yamauchi, F.; Nokihara, K. Antioxidant activity of designed peptides based on the antioxidative peptide derived from digests of a soybean peptide. J. Agric. Food Chem. 1996, 44, 2619–2623. [Google Scholar] [CrossRef]
- Chen, H.M.; Muramoto, K.; Yamauchi, F. Structural analysis of antioxidative peptides from soybean β-conglycinin. J. Agric. Food Chem. 1995, 43, 574–578. [Google Scholar] [CrossRef]
- Chen, Y.; Li, C.; Xue, J.; Kwok, L.Y.; Yang, J.; Zhang, H.; Menghe, B. Characterization of angiotensin-converting enzyme inhibitory activity of fermented milk produced by Lactobacillus helveticus. J. Dairy Sci. 2015, 98, 5113–5124. [Google Scholar] [CrossRef] [PubMed]





| Strain | ABTS (µmol TE/mL) | FRAP (µmol TE/mL) | ACEi Activity% |
|---|---|---|---|
| L. acidophilus (LA 14) | 12.07 ± 0.00 | 23.14 ± 0.00 | 36.81 ± 2.62 |
| L. helveticus (LH) | 42.07 ± 0.00 | 30.06 ± 0.00 | 74.37 ± 3.82 |
| Hydrolysate | ABTS (µmol TE/mL) | FRAP (µmol TE/mL) | ACEi Activity% * |
|---|---|---|---|
| SM-TCA | 17.00 ± 0.00 | 23.02 ± 0.00 | 81.77 ± 1.37 |
| BHI-TCA | 245.40 ± 0.00 | 29.55 ± 0.00 | 53.65 ± 1.82 |
| PW-TCA | 55.67 ± 0.00 | 9.58 ± 0.00 | 87.30 ± 1.59 |
| SM-Beads | 26.27 ± 0.00 | 20.84 ± 0.00 | 70.12 ± 3.81 |
| BHI-Beads | 261.40 ± 0.00 | 25.12 ± 0.00 | 49.59 ± 0.49 |
| PW-Beads | 64.87 ± 0.00 | 10.53 ± 0.00 | 97.75 ± 1.87 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ianni, F.; Altomare, A.A.; Cenci-Goga, B.T.; Blasi, F.; Grispoldi, L.; Regazzoni, L.; Cossignani, L. Chromatographic Characterization and In Vitro Bioactivity Evaluation of Lactobacillus helveticus Hydrolysates upon Fermentation of Different Substrates. Appl. Sci. 2021, 11, 811. https://0-doi-org.brum.beds.ac.uk/10.3390/app11020811
Ianni F, Altomare AA, Cenci-Goga BT, Blasi F, Grispoldi L, Regazzoni L, Cossignani L. Chromatographic Characterization and In Vitro Bioactivity Evaluation of Lactobacillus helveticus Hydrolysates upon Fermentation of Different Substrates. Applied Sciences. 2021; 11(2):811. https://0-doi-org.brum.beds.ac.uk/10.3390/app11020811
Chicago/Turabian StyleIanni, Federica, Alessandra Anna Altomare, Beniamino T. Cenci-Goga, Francesca Blasi, Luca Grispoldi, Luca Regazzoni, and Lina Cossignani. 2021. "Chromatographic Characterization and In Vitro Bioactivity Evaluation of Lactobacillus helveticus Hydrolysates upon Fermentation of Different Substrates" Applied Sciences 11, no. 2: 811. https://0-doi-org.brum.beds.ac.uk/10.3390/app11020811