Proteomic Characterization of Antibiotic Resistance, and Production of Antimicrobial and Virulence Factors in Streptococcus Species Associated with Bovine Mastitis. Could Enzybiotics Represent Novel Therapeutic Agents Against These Pathogens?
Abstract
:1. Introduction
2. Results
2.1. Streptococcus spp. Proteomics Data Repository
2.2. Proteins Involved in Bacterial Resistance to Antibiotics or Other Toxic Substances
2.3. Proteins Involved in Bacteriocin Production
2.4. Proteins Involved in Host Colonization and Immune Evasion
2.5. Proteins Involved in Bacterial Toxicity
2.6. Transporters Associated to Virulence Factors
3. Discussion
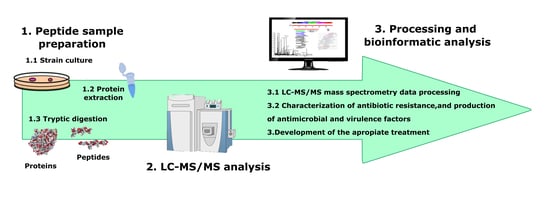
4. Materials And Methods
4.1. Bacterial Strains
4.2. Protein Extraction
4.3. Peptide Sample Preparation
4.4. Shotgun LC–ESI–MS/MS Analysis
4.5. LC–ESI–MS/MS Mass Spectrometry Data Processing
4.6. Determination of the Species Specificity of the Peptides Identified by LC–ESI–MS/MS
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Forsman, P.; Tilsala-Timisjärvi, A.; Alatossava, T. Identification of staphylococcal and streptococcal causes of bovine mastitis using 16S-23S rRNA spacer regions. Microbiology 1997, 143, 3491–3500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lopez-Sanchez, M.J.; Sauvage, E.; Da Cunha, V.; Clermont, D.; Ratsima Hariniaina, E.; Gonzalez-Zorn, B.; Poyart, C.; Rosinski-Chupin, I.; Glaser, P. The highly dynamic CRISPR1 system of Streptococcus agalactiae controls the diversity of its mobilome. Mol. Microbiol. 2012, 85, 1057–1071. [Google Scholar] [CrossRef] [PubMed]
- Richards, V.P.; Palmer, S.R.; Bitar, P.D.P.; Qin, X.; Weinstock, G.M.; Highlander, S.K.; Town, C.D.; Burne, R.A.; Stanhope, M.J. Phylogenomics and the dynamic genome evolution f the genus Streptococcus. Genome Biol. Evol. 2014, 6, 741–753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lundberg, Å.; Nyman, A.; Unnerstad, H.E.; Waller, K.P. Prevalence of bacterial genotypes and outcome of bovine clinical mastitis due to Streptococcus dysgalactiae and Streptococcus uberis. Acta Vet. Scand. 2014, 56, 80. [Google Scholar] [CrossRef] [Green Version]
- Dumke, J.; Hinse, D.; Vollmer, T.; Schulz, J.; Knabbe, C. Potential Transmission Pathways of Streptococcus gallolyticus subsp. gallolyticus. PLoS ONE 2015, 10. [Google Scholar] [CrossRef] [Green Version]
- Park, M.A.; Kwon, G.; Hwang, Y.; Jung, H.; Kim, D.; Park, J.; Kim, J. Genome Sequence of Streptococcus parauberis Strain KCTC11980, Isolated from Diseased Paralichthys olivaceus. Genome Announc. 2013, 1, 5–6. [Google Scholar] [CrossRef] [Green Version]
- Tian, X.Y.; Zheng, N.; Han, R.W.; Ho, H.; Wang, J.; Wang, Y.T.; Wang, S.Q.; Li, H.G.; Liu, H.W.; Yu, Z.N. Antimicrobial resistance and virulence genes of Streptococcus isolated from dairy cows with mastitis in China. Microb. Pathog. 2019, 131, 33–39. [Google Scholar] [CrossRef]
- Xiao, H.; Chen, X.; Chen, M.; Tang, S.; Zhao, X.; Huan, L. Bovicin HJ50, a novel lantibiotic produced by Streptococcus bovis HJ50. Microbiology 2004, 150, 103–108. [Google Scholar] [CrossRef]
- Begley, M.; Cotter, P.D.; Hill, C.; Ross, R.P. Identification of a novel two-peptide lantibiotic, lichenicidin, following rational genome mining for LanM proteins. Appl. Environ. Microbiol. 2009, 75, 5451–5460. [Google Scholar] [CrossRef] [Green Version]
- Maricic, N.; Anderson, E.S.; Opipari, A.M.E.; Yu, E.A.; Dawid, S. Characterization of a multipeptide lantibiotic locus in Streptococcus pneumoniae. MBio 2016, 7. [Google Scholar] [CrossRef] [Green Version]
- Nes, I.F.; Diep, D.B.; Holo, H. Bacteriocin diversity in Streptococcus and Enterococcus. J. Bacteriol. 2007, 189, 1189–1198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Antók, F.I.; Mayrhofer, R.; Marbach, H.; Masengesho, J.C.; Keinprecht, H.; Nyirimbuga, V.; Fischer, O.; Lepuschitz, S.; Ruppitsch, W.; Ehling-schulz, M.; et al. Characterization of antibiotic and biocide resistance genes and virulence factors of Staphylococcus species associated with bovine mastitis in Rwanda. Antibiotics 2020, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaczorek, E.; Małaczewska, J.; Wójcik, R.; Rękawek, W.; Siwicki, A.K. Phenotypic and genotypic antimicrobial susceptibility pattern of Streptococcus spp. isolated from cases of clinical mastitis in dairy cattle in Poland. J. Dairy Sci. 2017, 100, 6442–6453. [Google Scholar] [CrossRef] [PubMed]
- Villa, T.G.; Crespo, P.V. Enzybiotics: Antibiotic Enzymes as Drugs and Therapeutics; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2010. [Google Scholar]
- Dams, D.; Briers, Y. Enzybiotics: Enzyme-based antibacterials as therapeutics. In Advances in Experimental Medicine and Biology; Springer: New York, NY, USA, 2019; Volume 1148, pp. 233–253. [Google Scholar]
- Angelopoulou, A.; Warda, A.K.; Hill, C.; Ross, R.P. Non-antibiotic microbial solutions for bovine mastitis–live biotherapeutics, bacteriophage, and phage lysins. Crit. Rev. Microbiol. 2019, 45, 564–580. [Google Scholar] [CrossRef]
- Käppeli, N.; Morach, M.; Zurfluh, K.; Corti, S.; Nüesch-Inderbinen, M.; Stephan, R. Sequence types and antimicrobial resistance profiles of Streptococcus uberis isolated from bovine mastitis. Front. Vet. Sci. 2019, 6, 234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vélez, J.R.; Cameron, M.; Rodríguez-Lecompte, J.C.; Xia, F.; Heider, L.C.; Saab, M.; Trenton McClure, J.; Sánchez, J. Whole-genome sequence analysis of antimicrobial resistance genes in Streptococcus uberis and Streptococcus dysgalactiae isolates from Canadian dairy herds. Front. Vet. Sci. 2017, 4. [Google Scholar] [CrossRef]
- Tomazi, T.; de Souza Filho, A.F.; Heinemann, M.B.; dos Santos, M.V. Molecular characterization and antimicrobial susceptibility pattern of Streptococcus agalactiae isolated from clinical mastitis in dairy cattle. PLoS ONE 2018, 13, e01999561. [Google Scholar] [CrossRef]
- Carrera, M.; Böhme, K.; Gallardo, J.M.; Barros-Velázquez, J.; Cañas, B.; Calo-Mata, P. Characterization of foodborne strains of Staphylococcus aureus by shotgun proteomics: Functional networks, virulence factors and species-specific peptide biomarkers. Front. Microbiol. 2017, 8, 2458. [Google Scholar] [CrossRef] [Green Version]
- Calo-mata, P.; Carrera, M.; Böhme, K.; Caamaño-Antelo, S.; Gallardo, J.M.; Barros-Velázquez, J.; Cañas, B. Novel peptide biomarker discovery for detection and identification of bacterial pathogens by LC-ESI-MS/MS. J. Anal. Bioanal. Tech. 2016, 7, 1–9. [Google Scholar]
- Pfrunder, S.; Grossmann, J.; Hunziker, P.; Brunisholz, R.; Gekenidis, M.; Drissner, D. Bacillus cereus group-type strain-specific diagnostic peptides. J. Proteome Res. 2016, 15, 3098–3107. [Google Scholar] [CrossRef] [Green Version]
- Hynes, W.; Sloan, M. Secreted extracellular virulence factors. In Streptococcus Pyogenes: Basic Biology to Clinical Manifestations; Ferretti, J.J., Stevens, D.L., Fischetti, V.A., Eds.; University of Oklahoma Health Sciences Center: Oklahoma City, OK, USA, 2016; pp. 1–40. [Google Scholar]
- Golińska, E.; Van der Linden, M.; Więcek, G.; Mikołajczyk, D.; Machul, A.; Samet, A.; Piórkowska, A.; Dorycka, M.; Heczko, P.B.; Strus, M. Virulence factors of Streptococcus pyogenes strains from women in peri-labor with invasive infections. Eur. J. Clin. Microbiol. Infect. Dis. 2016, 35, 747–754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Grove, A. Quick guide MarR family transcription factors. Curr. Biol. 2013, 23, R142–R143. [Google Scholar] [CrossRef] [Green Version]
- Zapun, A.; Contreras-martel, C.; Vernet, T. Penicillin-binding proteins and b -lactam resistance. FEMS Microbiol. Lett. 2008, 32, 361–385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- López-pelegrín, M.; Cerdà-costa, N.; Martínez-jiménez, F.; Cintas-pedrola, A.; Canals, A.; Peinado, J.R.; Marti-renom, M.A.; López-otín, C.; Arolas, J.L.; Gomis-rüth, F.X. A Novel family of soluble minimal scaffolds provides structural insight into the catalytic domains of integral membrane metallopeptidases. J. Biol. Chem. 2013, 288, 21279–21294. [Google Scholar] [CrossRef] [Green Version]
- Kahmann, J.D.; Sass, H.; Allan, M.G.; Seto, H.; Thompson, C.J.; Grzesiek, S. Structural basis for antibiotic recognition by the TipA class of multidrug-resistance transcriptional regulators. EMBO J. 2003, 22, 1824–1834. [Google Scholar] [CrossRef] [Green Version]
- Evans, J.J.; Bolz, D.D. Regulation of virulence and antibiotic resistance in Gram-positive microbes in response to cell wall-active antibiotics. Curr. Opin. Infect. Dis. 2019, 32, 217–222. [Google Scholar] [CrossRef]
- Kehrenberg, C.; Catry, B.; Haesebrouck, F.; De Kruif, A.; Schwarz, S. Novel Spectinomycin/Streptomycin Resistance Gene, aadA14, from Pasteurella multocida. Antimicrob. Agents Chemother. 2005, 49, 3046–3049. [Google Scholar] [CrossRef] [Green Version]
- Dowson, C.G.; Hutchison, A.; Brannigan, J.A.; George, R.C.; Hansman, D.; Linares, J.; Tomasz, A.; Smith, J.M.; Spratt, B.G. Horizontal transfer of penicillin-binding protein genes in penicillin-resistant clinical isolates of Streptococcus pneumoniae. Proc. Natl. Acad. Sci. USA 1989, 86, 8842–8846. [Google Scholar] [CrossRef] [Green Version]
- Suprenant, K.A.; Bloom, N.; Fang, J.; Lushington, G. The major vault protein is related to the toxic anion resistance protein (TelA) family. J. Exp. Biol. 2007, 210, 946–955. [Google Scholar] [CrossRef] [Green Version]
- Hobman, J.L.; Wilkie, J.; Brown, N.L. A design for life: Prokaryotic metal-binding MerR family regulators. BioMetals 2005, 18, 429–436. [Google Scholar] [CrossRef] [PubMed]
- Dash, H.R.; Sahu, M.; Mallick, B.; Das, S. Functional efficiency of MerA protein among diverse mercury resistant bacteria for efficient use in bioremediation of inorganic mercury. Biochimie 2017, 142, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Bateman, A.; Bycroft, M. The structure of a LysM domain from E. coli membrane-bound lytic murein transglycosylase D (MltD). J. Mol. Biol. 2000, 299, 1113–1119. [Google Scholar] [CrossRef] [PubMed]
- Wisniewski, J.A.; Rood, J.I. The Tcp conjugation system of Clostridium perfringens. Plasmid 2017, 91, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Kajfasz, J.K.; Martinez, A.R.; Rivera-ramos, I.; Abranches, J.; Koo, H.; Quivey, R.G. Role of Clp Proteins in expression of virulence properties of Streptococcus mutans. J. Bacteriol. 2009, 191, 2060–2068. [Google Scholar] [CrossRef] [Green Version]
- Kwon, H.; Ogunniyi, A.D.; Choi, M.; Pyo, S.; Rhee, D.; Paton, J.C. The ClpP Protease of Streptococcus pneumoniae modulates virulence gene expression and protects against fatal pneumococcal challenge. Infect. Immun. 2004, 72, 5646–5653. [Google Scholar] [CrossRef] [Green Version]
- Manco, S.; Hernon, F.; Yesilkaya, H.; Paton, J.C.; Andrew, P.W.; Kadioglu, A. Pneumococcal Neuraminidases A and B both have essential roles during infection of the respiratory tract and sepsis. Infect. Immun. 2006, 74, 4014–4020. [Google Scholar] [CrossRef] [Green Version]
- Xiao, K.; Wang, X.; Yu, H. Comparative studies of catalytic pathways for Streptococcus pneumoniae sialidases NanA, NanB and NanC. Sci. Rep. Nat. 2019, 9, 1–13. [Google Scholar] [CrossRef]
- Galán-Bartual, S.; Pérez, I.; Hermoso, J.A.; Garcıa, P. Structure and Function of Choline-Binding Proteins; Academic Press: London, UK; San Diego, CA, USA; Waltham, MA, USA; Oxford, UK, 2015; pp. 207–230. [Google Scholar]
- Eckhard, U.; Bandukwala, H.; Mans, M.J.; Marino, G.; Holyoak, T.; Charles, T.C.; Austin, J.; Overall, C.M.; Cheng, J.; Wallace, I.; et al. Discovery of a proteolytic flagellin family in diverse bacterial phyla that assembles enzymatically active flagella. Nat. Commun. 2017, 8, 521. [Google Scholar] [CrossRef]
- Jacobitz, A.W.; Kattke, M.D.; Wereszczynski, J.; Clubb, R.T. Sortase Transpeptidases: Structural Biology and Catalytic Mechanism. In Advances in Protein Chemistry and Structural Biology; Academic Press Inc.: London, UK; San Diego, CA, USA; Waltham, MA, USA; Oxford, UK, 2017; Volume 109, pp. 223–264. [Google Scholar]
- Ribardo, D.A.; Lambert, T.J.; Mciver, K.S. Role of Streptococcus pyogenes two-component response regulators in the temporal control of Mga and the Mga-regulated virulence gene emm. Infect. Immun. 2004, 72, 3668–3673. [Google Scholar] [CrossRef] [Green Version]
- Benachour, A.; Ladjouzi, R.; Thorpe, S.; Courtin, P.; Prajsnar, T.K.; Foster, S.J. The lysozyme-induced peptidoglycan N -acetylglucosamine deacetylase PgdA (EF1843) is required for Enterococcus faecalis. J. Bacteriol. 2012, 194, 6066–6073. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vollmer, W.; Tomasz, A. Peptidoglycan N -acetylglucosamine deacetylase, a putative virulence factor in Streptococcus pneumoniae. Infect. Immun. 2002, 70, 7176–7178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Broxton, C.N.; Culotta, V.C. SOD Enzymes and Microbial Pathogens: Surviving the Oxidative Storm of Infection. PLoS Pathog. 2016, 12, 8–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cianciotto, N.P.; White, R.C. Expanding role of type II secretion in bacterial pathogenesis and beyond. Infect. Immun. 2017, 85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Warne, B.; Harkins, C.P.; Harris, S.R.; Vatsiou, A.; Stanley-Wall, N.; Parkhill, J.; Peacock, S.J.; Palmer, T.; Holden, M.T.G. The Ess/Type VII secretion system of Staphylococcus aureus shows unexpected genetic diversity. BMC Genom. 2016, 17, 222. [Google Scholar] [CrossRef] [Green Version]
- Paton, J.C.; Trappetti, C. Streptococcus pneumoniae capsular polysaccharide. In Gram-Positive Pathogens; ASM Press: Washington, DC, USA, 2019; pp. 304–315. [Google Scholar]
- Ali, L.; Spiess, M.; Wobser, D.; Rodriguez, M.; Blum, H.E.; Saknç, T. Identification and functional characterization of the putative polysaccharide biosynthesis protein (CapD) of Enterococcus faecium U0317. Infect. Genet. Evol. 2016, 37, 215–224. [Google Scholar] [CrossRef]
- Epler Barbercheck, C.R.; Bullitt, E.; Andersson, M. Bacterial adhesion pili. In Subcellular Biochemistry; Springer: New York, NY, USA, 2018; Volume 87, pp. 1–18. [Google Scholar]
- Angelov, A.; Bergen, P.; Nadler, F.; Hornburg, P.; Lichev, A.; Ãœbelacker, M.; Pachl, F.; Kuster, B.; Liebl, W. Novel Flp pilus biogenesis-dependent natural transformation. Front. Microbiol. 2015, 6, 84. [Google Scholar] [CrossRef] [Green Version]
- Pellecchia, M.; Sebbel, P.; Hermanns, U.; Wüthrich, K.; Glockshuber, R. Pilus chaperone FimC-adhesin FimH interactions mapped by TROSY-NMR. Nat. Struct. Biol. 1999, 6, 336–339. [Google Scholar]
- Demuth, D.R.; Lammey, M.S.; Huck, M.; Lally, E.T.; Malamud, D. Comparison of Streptococcus mutans and Streptococcus sanguis receptors for human salivary agglutinin. Microb. Pathog. 1990, 9, 199–211. [Google Scholar] [CrossRef]
- Narasaki, R.; Kuribayashi, H.; Shimizu, K.; Imamura, D.; Sato, T.; Hasumi, K. Bacillolysin MA, a novel bacterial metalloproteinase that produces angiostatin-like fragments from plasminogen and activates protease zymogens in the coagulation and fibrinolysis systems. J. Biol. Chem. 2005, 280, 14278–14287. [Google Scholar] [CrossRef] [Green Version]
- Rivière, G.; Peng, E.Q.; Brotgandel, A.; Andring, J.T.; Lakshmanan, R.V.; Agbandje-McKenna, M.; McKenna, R.; Brady, L.J.; Long, J.R. Characterization of an intermolecular quaternary interaction between discrete segments of the Streptococcus mutans adhesin P1 by NMR spectroscopy. FEBS J. 2019. [Google Scholar] [CrossRef] [PubMed]
- Angel, C.S.; Ruzek, M.; Hostetter, M.K. Degradation of C3 by Streptococcus pneumoniae. J. Infect. Dis. 1994, 170, 600–608. [Google Scholar] [CrossRef] [PubMed]
- Sharma-Kuinkel, B.K.; Mann, E.E.; Ahn, J.S.; Kuechenmeister, L.J.; Dunman, P.M.; Bayles, K.W. The Staphylococcus aureus LytSR two-component regulatory system affects biofilm formation. J. Bacteriol. 2009, 191, 4767–4775. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petersen, I.; Schlüter, R.; Hoff, K.J.; Liebscher, V.; Bange, G.; Riedel, K.; Pané-Farré, J. Non-invasive and label-free 3D-visualization shows in vivo oligomerization of the staphylococcal alkaline shock protein 23 (Asp23). Sci. Rep. 2020, 10, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Holden, V.I.; Bachman, M.A. Diverging roles of bacterial siderophores during infection. Metallomics 2015, 7, 986–995. [Google Scholar] [CrossRef]
- Heather, Z.; Holden, M.T.G.; Steward, K.F.; Parkhill, J.; Song, L.; Challis, G.L.; Robinson, C.; Davis-Poynter, N.; Waller, A.S. A novel streptococcal integrative conjugative element involved in iron acquisition. Mol. Microbiol. 2008, 70, 1274–1292. [Google Scholar] [CrossRef] [Green Version]
- Nieto, C.; Cherny, I.; Khoo, S.K.; Garcı, M.; Lacoba, D.; Chan, W.T. The yefM-yoeb toxin-antitoxin systems of Escherichia coli and Streptococcus pneumoniae: Functional and Structural Correlation. J. Bacteriol. 2007, 189, 1266–1278. [Google Scholar] [CrossRef] [Green Version]
- Chan, W.T.; Nieto, C.; Harikrishna, J.A.; Khoo, S.K.; Othman, R.Y.; Espinosa, M.; Yeo, C.C. Genetic regulation of the yefM-yoeB toxin-antitoxin locus of Streptococcus pneumoniae. J. Bacteriol. 2011, 193, 4612–4625. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.; Wang, X.; Ma, Q.; Zhang, X.S.; Wood, T.K. Toxin-antitoxin systems in Escherichia coli influence biofilm formation through YjgK (TabA) and fimbriae. J. Bacteriol. 2009, 191, 1258–1267. [Google Scholar] [CrossRef] [Green Version]
- Commons, R.; Rogers, S.; Gooding, T.; Danchin, M.; Carapetis, J.; Robins-Browne, R.; Curtis, N. Superantigen. Superantigen genes in group A streptococcal isolates and their relationship with emm types. J. Med. Microbiol. 2008, 57, 1238–1246. [Google Scholar] [CrossRef]
- Lappin, E.; Ferguson, A.J. Gram-positive toxic shock syndromes. Lancet Infect. Dis. 2009, 9, 281–290. [Google Scholar] [CrossRef]
- Proft, T.; Fraser, J.D. Streptococcal superantigens: Biological properties and potential role in disease. In Streptococcus Pyogenes: Basic Biology to Clinical Manifestations; Ferretti, J.J., Stevens, D.L., Fischetti, V.A., Eds.; University of Oklahoma Health Sciences Center: Oklahoma City, OK, USA, 2016. [Google Scholar]
- Peschel, A.; Otto, M. Phenol-soluble modulins and staphylococcal infection. Nat. Rev. Microbiol. 2013, 11, 667–673. [Google Scholar] [CrossRef] [PubMed]
- Page, R.; Peti, W. Toxin-antitoxin systems in bacterial growth arrest and persistence. Nat. Chem. Biol. 2016, 12, 208–214. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K.J.; Song, S.; Mason, K.; Pinkett, H.W. Selective substrate uptake: The role of ATP-binding cassette (ABC) importers in pathogenesis. Biochim. Biophys. Acta 2018, 1860, 868–877. [Google Scholar] [CrossRef]
- Price, C.T.; Bukka, A.; Cynamon, M.; Graham, J.E.; Al, P.E.T.; Acteriol, J.B. Glycine betaine uptake by the ProXVWZ ABC transporter contributes to the ability of Mycobacterium tuberculosis to initiate growth in human macrophages. J. Bacteriol. 2008, 190, 3955–3961. [Google Scholar] [CrossRef] [Green Version]
- Finkenwirth, F.; Sippach, M.; Pecina, S.N.; Gäde, M.; Ruta, J.; Ricke, A.; Bondarenko, E.; Klare, J.P.; Zinke, M.; Lange, S.; et al. Dynamic interactions of CbiN and CbiM trigger activity of a cobalt energy-coupling-factor transporter. Biochim. Biophys. Acta 2020, 1862, 183114. [Google Scholar] [CrossRef]
- Lau, C.K.Y.; Krewulak, K.D.; Vogel, H.J. Bacterial ferrous iron transport: The Feo system. FEMS Microbiol. Rev. 2020, 40, 273–298. [Google Scholar] [CrossRef]
- Ma, J.; Liu, J.; Zhang, Y.; Wang, D.; Liu, R.; Liu, G.; Yao, H.; Pan, Z. Bacitracin resistance and enhanced virulence of Streptococcus suis via a novel efflux pump. BMC Vet. Res. 2019, 15, 377. [Google Scholar] [CrossRef]
- Orelle, C.; Mathieu, K.; Jault, J.M. Multidrug ABC transporters in bacteria. Res. Microbiol. 2019, 170, 381–391. [Google Scholar] [CrossRef]
- Greene, N.P.; Kaplan, E.; Crow, A.; Koronakis, V. Antibiotic resistance mediated by the MacB ABC transporter family: A structural and functional perspective. Front. Microbiol. 2018, 9, 950. [Google Scholar] [CrossRef] [Green Version]
- Bull, P.C.; Cox, D.W. Wilson disease and Menkes disease: New handles on heavy-metal transport. Trends Genet. 1994, 10, 246–252. [Google Scholar] [CrossRef]
- Kumar, S.; Mukherjee, M.M.; Varela, M.F. Modulation of bacterial multidrug resistance efflux pumps of the major facilitator superfamily. Int. J. Bacteriol. 2013, 2013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Omote, H.; Hiasa, M.; Matsumoto, T.; Otsuka, M.; Moriyama, Y. The MATE proteins as fundamental transporters of metabolic and xenobiotic organic cations. Trends Pharmacol. Sci. 2006, 27, 587–593. [Google Scholar] [CrossRef] [PubMed]
- Böhme, K.; Morandi, S.; Cremonesi, P.; Fernández No, I.C.; Barros-Velázquez, J.; Castiglioni, B.; Brasca, M.; Cañas, B.; Calo-Mata, P. Characterization of Staphylococcus aureus strains isolated from Italian dairy products by MALDI-TOF mass fingerprinting. Electrophoresis 2012, 33, 2355–2364. [Google Scholar] [CrossRef]
- Quintela-Baluja, M.; Böhme, K.; Fernández-No, I.C.; Alnakip, M.E.; Caamano, S.; Barros-Velázques, J.; Calo-mata, P. MALDI-TOF Mass spectrometry, a rapid and reliable method for the identification of bacterial species in food-microbiology laboratories. In Novel Food Preservation and Microbial Assessment Techniques; Boziaris, L.S., Ed.; CRC Press: Boca Raton, FL, USA, 2014; pp. 353–385. [Google Scholar]
- Böhme, K.; Fernández-No, I.C.; Barros-Velázquez, J.; Gallardo, J.M.; Calo-Mata, P.; Cañas, B. Species differentiation of seafood spoilage and pathogenic gram-negative bacteria by MALDI-TOF mass fingerprinting. J. Proteome Res. 2010, 9, 3169–3183. [Google Scholar] [CrossRef]
- Ganaie, M.Y.; Qureshi, S.; Kashoo, Z.; Wani, S.A.; Hussain, M.I.; Kumar, R.; Maqbool, R.; Sikander, P.; Banday, M.S.; Malla, W.A.; et al. Isolation and characterization of two lytic bacteriophages against Staphylococcus aureus from India: Newer therapeutic agents against Bovine mastitis. Vet. Res. Commun. 2018, 42, 289–295. [Google Scholar] [CrossRef]
- Geng, H.; Zou, W.; Zhang, M.; Xu, L.; Liu, F.; Li, X.; Wang, L.; Xu, Y. Evaluation of phage therapy in the treatment of Staphylococcus aureus-induced mastitis in mice. Folia Microbiol. 2019, 65, 339–351. [Google Scholar] [CrossRef]
- Heselpoth, R.D.; Swift, S.M.; Linden, S.B.; Mitchell, M.S.; Nelson, D.C. Enzybiotics: Endolysins and Bacteriocins. In Bacteriophages; Harper, D.R., Abedon, S.T., Burrowes, B.H., McConville, M.L., Eds.; Springer: Cham, Switzerland, 2018; pp. 1–42. [Google Scholar]
- Fan, J.; Zeng, Z.; Mai, K.; Yang, Y.; Feng, J.; Bai, Y.; Sun, B.; Xie, Q.; Tong, Y.; Ma, J. Preliminary treatment of bovine mastitis caused by Staphylococcus aureus, with trx-SA1, recombinant endolysin of S. aureus bacteriophage IME-SA1. Vet. Microbiol. 2016, 191, 65–71. [Google Scholar] [CrossRef]
- Schmelcher, M.; Powell, A.M.; Camp, M.J.; Pohl, C.S.; Donovan, D.M. Synergistic streptococcal phage λSA2 and B30 endolysins kill streptococci in cow milk and in a mouse model of mastitis. Appl. Microbiol. Biotechnol. 2015, 99, 8475–8486. [Google Scholar] [CrossRef]
- Lin, D.M.; Koskella, B.; Lin, H.C. Phage therapy: An alternative to antibiotics in the age of multi-drug resistance. World J. Gastrointest. Pharmacol. Ther. 2017, 8, 162. [Google Scholar] [CrossRef]
- Zhang, L.; Gao, J.; Barkema, H.W.; Ali, T.; Liu, G.; Deng, Y.; Naushad, S.; Kastelic, J.P.; Han, B. Virulence gene profiles: Alpha-hemolysin and clonal diversity in Staphylococcus aureus isolates from bovine clinical mastitis in China. BMC Vet. Res. 2018, 14, 63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Collado, R.; Prenafeta, A.; González-González, L.; Pérez-Pons, J.A.; Sitjà, M. Probing vaccine antigens against bovine mastitis caused by Streptococcus uberis. Vaccine 2016, 34, 3848–3854. [Google Scholar] [CrossRef] [PubMed]
- Perrig, M.S.; Veaute, C.; Renna, M.S.; Pujato, N.; Calvinho, L.; Marcipar, I.; Barbagelata, M.S. Assessment of the potential utility of different regions of Streptococcus uberis adhesion molecule (SUAM) for mastitis subunit vaccine development. Microb. Pathog. 2017, 105, 273–279. [Google Scholar] [CrossRef]
- Aranha, M.P.; Penfound, T.A.; Spencer, J.A.; Agarwal, R.; Baudry, J.; Dale, J.B.; Smith, J.C.; Cresswell, P. Structure-based group A streptococcal vaccine design: Helical wheel homology predicts antibody cross-reactivity among streptococcal M protein-derived peptides. J. Biol. Chem. 2020, 295, 3826–3836. [Google Scholar] [CrossRef] [PubMed]
- Pastural, É.; McNeil, S.A.; MacKinnon-Cameron, D.; Ye, L.; Langley, J.M.; Stewart, R.; Martin, L.H.; Hurley, G.J.; Salehi, S.; Penfound, T.A.; et al. Safety and immunogenicity of a 30-valent M protein-based group a streptococcal vaccine in healthy adult volunteers: A randomized, controlled phase I study. Vaccine 2020, 38, 1384–1392. [Google Scholar] [CrossRef] [PubMed]
- Courtney, H.S.; Niedermeyer, S.E.; Penfound, T.A.; Hohn, C.M.; Greeley, A.; Dale, J.B. Trivalent M-related protein as a component of next generation group A streptococcal vaccines. Clin. Exp. Vaccine Res. 2017, 6, 45–49. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Zhao, Y.; Wang, G.; Feng, S.; Guo, Z.; Gu, G. Group A Streptococcus Cell Wall Oligosaccharide-Streptococcal C5a Peptidase Conjugates as Effective Antibacterial Vaccines. ACS Infect. Dis. 2020, 6, 281–290. [Google Scholar] [CrossRef]
- Raynes, J.M.; Young, P.G.; Proft, T.; Williamson, D.A.; Baker, E.N.; Moreland, N.J. Protein adhesins as vaccine antigens for Group A Streptococcus. Pathog. Dis. 2018, 76. [Google Scholar] [CrossRef]
- Dale, J.B.; Penfound, T.A.; Chiang, E.Y.; Walton, W.J. New 30-valent M protein-based vaccine evokes cross-opsonic antibodies against non-vaccine serotypes of group A streptococci. Vaccine 2011, 29, 8175–8178. [Google Scholar] [CrossRef] [Green Version]
- Carrera, M.; Cañas, B.; Gallardo, J.M. The sarcoplasmic fish proteome: Pathways, metabolic networks and potential bioactive peptides for nutritional inferences. J. Proteom. 2013, 78, 211–220. [Google Scholar] [CrossRef]
- Käll, L.; Canterbury, J.D.; Weston, J.; Noble, W.S.; MacCoss, M.J. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods 2007, 4, 923–925. [Google Scholar] [CrossRef] [PubMed]
| Function | Strain | Protein | Peptide | Identity by BLASTP |
|---|---|---|---|---|
| Toxins | ST1 | Toxin RelE | LLATISM*IQEQGVLIAQRM*EWVKK | Streptococcus suis |
| ST3 | Antitoxin RelB | VFKENNLNTAQALNLFLKNVAETGQLNLK | Streptococcus gallolyticus | |
| ST12 | Antitoxin YefM | NTYLSQKVLRGM*AK | Streptococcus suis | |
| ST2 | Toxin YoeB | LIYM*M*DGDNVAFLSFKDHY | Streptococcus mitis | |
| ST11 and ST12 | Pyrogenic exotoxin SpeK | NIYAPRYDEDEILDNR | Streptococcus dysgalactiae subsp. dysgalactiae, Streptococcus dysgalactiae | |
| ST9 | Doc toxin | LYPTLFDKATILFVQLVKK | Streptococcus sobrinus Streptococcus downei | |
| Antibiotic resistance | ST3 | MarR family transcriptional regulator | M*DYQRINDYLTSIFNNVLVIEEM*SLRGSR | Streptococcus spp. |
| ST4 | MarR family transcriptional regulator | FNRFILAFEQLKK | Streptococcus oralis | |
| ST2 | MarR family transcriptional regulator | EM*QQYVDLQGAYLALVKEEFAKAGLLPLK | Streptococcus downei MFe28 | |
| ST9 | MurM protein | QSLQRYLSEFRGFLDK | Streptococcus equi | |
| ST8 | Beta-lactamase class A | FSITDVLVNSKKELVFQIDDK | Streptococcus suis | |
| ST6 | Beta-lactamase class A | LVPDQPIQITGFYVNEEEVPIFKLKNGQFVIADK | Streptococcus sanguinis | |
| ST3 | Cell wall-active antibiotics response protein | DTIHLERVILSNHDNVIILRK | Streptococcus pseudopneumoniae, Streptococcus sp. HMSC061D10, Streptococcus sp. SK140 | |
| ST10 | Streptomycin adenylyltransferase | M*RTETDM*FDVILQTAKVLQVDAVAM*SGSR | Streptococcus cristatus | |
| ST12 | Penicillin binding protein | VQESAQNAGDTIGRAVK | Streptococcus gallolyticus, Streptococcus macedonicus, Streptococcus pasteurianus | |
| ST14 | Glyoxalase/Bleomycin resistance protein | M*ITSLYPVLM*C*ENLEATANFFIENFQFR | Streptococcus sp. DD11 | |
| ST1 | M56 peptidase | FSHGQTAHETIVNAKDGKLVK | Streptococcus sanguinis | |
| Resistance to toxic substances | ST4 | TelA protein | DSLQEFYFDSKSIEQKM*DGM*AAAVVK | Streptococcus iniae |
| ST1 | MerR family transcriptional regulator | LEDHLLDLKAK | Streptococcus agalactiae, Streptococcus halotolerans, Streptococcus thoraltensis, Streptococcus acidominimus | |
| Colonization and immune evasion | ST1 | N-acetylmuramoyl-L-alanine amidase | M*KKVILASTVALSILGFTQATVQAQENNAESVR | Streptococcus mitis |
| ST7 | N-acetylmuramoyl-L-alanine amidase | LVEIAFIDNNSDM*ATYEANK | Streptococcus dysgalactiae, Streptococcus urinalis, Streptococcus porcinus, Streptococcus agalactiae, Streptococcus pluranimalium, Streptococcus suis | |
| ST8 | Lysin | AGAIFVKREASHDYGHTGVVIK | Streptococcus phocae | |
| ST10 | Lysozyme | LIIFLLVFLFAFQTYR | Streptococcus henryi | |
| ST4 | Lysozyme M1 (1,4-beta-N-acetylmuramidase) | LNPM*IVVVFFLSFFALIFITGVTGNTVNK | Streptococcus suis | |
| ST3 | CLpX ATPases | EENDVDLQKSNILM*IGPTGSGKTFLAQTLAR | Streptococcus vestibularis | |
| ST5 | CLpX ATPases | SIIEETM*LDVM*FEVPSQENVKLIRITK | Streptococcus pneumoniae | |
| ST5 | CLp ATPases | WIGDAQKRTK | Streptococcus agalactiae, Streptococcus canis, Streptococcus equi, Streptococcus castoreus, Streptococcus dysgalactiae | |
| ST9 | CLp ATPases | RTIQDHIEDAITDYYLEHPK | Streptococcus cristatus, Streptococcus gordonii | |
| ST8 | CLp ATPases | ENLLQIVELM*LADVNKRLSSNNIHLDVTDK | Streptococcus pneumoniae, Streptococcus mitis | |
| ST8 | CLpC ATPases | EDVVKLIGNRATR | Streptococcus sinensis, Streptococcus anginosus | |
| ST14 | CLpX ATPases | NNPVLVGDAGVGKTVLALGLAQR | Streptococcus suis, Streptococcus pneumoniae | |
| ST3 | CLp ATPases | IM*VQPLIAHLAEKNISLK | Streptococcus macacae | |
| ST7 | CLp ATPases | ETIKAIHDLRKPK | Streptococcus castoreus, Streptococcus ictaluri | |
| ST6 | Neuraminidase A | SLVLPKLPGQVSLIGSNKQGVVDLNNK | Streptococcus sp. HMSC074B11, Streptococcus pseudopneumoniae, Streptococcus sp. HPH0090, Streptococcus sp. oral taxon 431, Streptococcus mitis, Streptococcus sp. UMB0029, Streptococcus sp. LQJ-218, Streptococcus infantis | |
| ST6 | Sialidase B | NAPYLGPGRGIIESSTGRILIPSYTGK | Streptococcus pneumoniae, Streptococcus mitis, Streptococcus pseudopneumoniae, Streptococcus infantis | |
| ST13 | Sialidase A | VPLVTSGDYSGSPINM*DM*ALVQDTSSKTK | Streptococcus agalactiae | |
| ST14 | Sialidase A | VPTLQLANGKTARFM*TQYDTK | Streptococcus pneumoniae, Streptococcus oralis | |
| ST3 | Sialidase A | EDVETNTSNGQRVDLSSELDKLK | Streptococcus pneumoniae | |
| ST10 | Choline binding protein (Cbp) | TGWVKDKGTWYYLDK | Streptococcus pneumoniae | |
| ST2 | Choline binding protein (Cbp) | EGSTWYYLKGSGAM*ATGWATANGQWSYFEK | Streptococcus mitis | |
| ST7 | PspA | TEQVLLTEAVQQVQR | Streptococcus gordonii, Streptococcus cristatus | |
| ST4 | PspA | DLDAADKALEAAQAELKAR | Streptococcus mitis | |
| ST4 | Ig A1 protease | GTESEAAKPAPKEAGTTAGNEVK | Streptococcus pneumoniae | |
| ST2 | Ig A1 protease | NNDKYYAIYNLK | Streptococcus sp. 596553, Streptococcus pneumoniae | |
| ST2 | Ig A1 protease | KKVM*GLLLIGSM*GQSLLLSIDAAALQNIELR | Streptococcus spp. | |
| ST13 | Sortase A | AKVGM*TIYLTDKSM*IYTYK | Streptococcus gallolyticus, Streptococcus macedonicus, Streptococcus pasteurianus, Streptococcus henryi | |
| ST2 | Sortase B | NFLIGQQSNHYQVSKVSKK | Streptococcus macedonicus, Streptococcus gallolyticus, Streptococcus pasteurianus, Streptococcus lutetiensis, | |
| ST4 | Sortase A | YYYEAAFLIIVPENTAFYK | Streptococcus azizi, Streptococcus acidominimus | |
| ST6 and ST13 | C5A peptidase | EDISGEEASAPQTSPQESPVEPEEVTRGR | Streptococcus suis | |
| ST2 | C5A peptidase | YPDKSPAEISELVKALIM*STAKPHINK | Streptococcus anginosus | |
| ST13 | M protein | LM*EERARHVDLIDNIR | Streptococcus pyogenes | |
| ST1 | M Protein | SVAVAVAVLGAAFANQTEVK | Streptococcus pyogenes | |
| ST1 | M Protein | AEAVSRSNSEQNNLEKR | Streptococcus pyogenes | |
| ST14 | M Protein | IVAVALTVVGAGFANQTEVK | Streptococcus pyogenes | |
| ST11 | M Protein | YVEKSYHLLSDFIDQISSTYNFKIDNK | Streptococcus cristatus | |
| ST9 | Mga protein | KVLLTFFLDKR | Streptococcus pseudoporcinus | |
| ST5 | O-acetylase OafA | IVPPLVM*M*ILLIIPFTFLVR | Streptococcus henryi | |
| ST10 | Superoxide dismutase | FGSGWAWLVVNPDGKLEVM*STANQDTPISEGK | Streptococcus anginosus, Streptococcus anginosus subsp. anginosus, Streptococcus constellatus subsp. constellatus, Streptococcus sp. 8400103 | |
| ST14 | Superoxide dismutase | FGSGWAWLVVNKDGKLEVTSTANQDTPLSEGK | Streptococcus infantarius, Streptococcus equinus | |
| ST6 | Peptidoglycane-N-acetylglucosamine deacetylase | DAELYQTYFAQK | Streptococcus oralis | |
| ST6 | CpsB | KGM*FETPEEKIAENFLQIR | Streptococcus pneumoniae | |
| ST1 | CpsC | EIILSQDVLEKVATDLKLELPPK | Streptococcus sp. 1643, Streptococcus oralis | |
| ST5 | CpsC | EIIISQDVLEEVVSDLKLDLTPK | Streptococcus pneumoniae | |
| ST13 | CapD protein | KLTDYVIDLVEILNK | Streptococcus pneumoniae, mitis, Streptococcus pseudopneumoniae, Streptococcus oralis, Streptococcus australis, Streptococcus sp. M334 | |
| ST3 | Accessory pilus subunit | NNVKTYLLKIK | Streptococcus suis | |
| ST8 | Pilin protein FimC | SRFGDAADKAASLSAK | Streptococcus sanguinis | |
| ST12 | Agglutinin receptor | TVETIQSTNEQAVADYLTKKTK | Streptococcus suis, Streptococcus agalactiae | |
| ST3 | Agglutinin receptor | VESAVSLAKEAGLTVK | Streptococcus mitis | |
| ST7 | Agglutinin receptor | TIDPSVHQYGQQELDALVK | Streptococcus oralis, Streptococcus sp. CM6, Streptococcus sp. SR1 | |
| ST9 | Agglutinin receptor | TTSLM*FEDYLPAGYLFDLEKTLAENGDYEVTFDASK | Streptococcus canis FSL Z3-227 | |
| ST8 | adhesin P1/ Cell surface antigen I/II | ADYEAKLAKYQADLAK | Streptococcus mutans, Streptococcus intermedius, Streptococcus anginosus | |
| ST7 | CppA protein | NLFQGRENFIPK | Streptococcus anginosus | |
| ST9 | Transposase TcpC | TLEQFLDGYVSRYFTYDSQAGSSDENISK | Streptococcus pneumoniae, Streptococcus oralis, Streptococcus sp. HMSC056C01, Streptococcus sp. SK140, Streptococcus infantis SK1302 | |
| ST5 | LytR family transcriptional regulator | AHTVQIITEEASFNM*VQNLSNLENQYGETLM*R | Streptococcus oralis | |
| ST2 | Asp23 protein | SGLSGGFSAVQEKVGEGVEAVKDAASSNENTR | Streptococcus cristatus | |
| ST12 | Asp23protein | KM*TDLDVIEVNVKVVDIK | Streptococcus phocae, Streptococcus canis, Streptococcus ictaluri, Streptococcus pyogenes, Streptococcus dysgalactiae, Streptococcus dysgalactiae subsp. equisimilis, Streptococcus dysgalactiae subsp. dysgalactiae, Streptococcus dysgalactiae subsp. equisimilis SK1249 | |
| ST14 | Asp23 protein | ATEDGSIAVDVYTVLSYGTKISEVSKNIQER | Streptococcus infantis, Streptococcus oralis, Streptococcus mitis | |
| ST2 | Type VII secretion protein EsaA | NSDVSTALSNIWFEAIDSNLKK | Streptococcus oralis | |
| ST2 | Type VII secretion protein EssB | LRLALNLLDLEQALSLPVTFFLHPENLFITK | Streptococcus pantholopis | |
| ST8 | Type VII secretion protein EssB | LEFVREDNQISVQISSSGYRR | Streptococcus sp., Streptococcus mitis | |
| ST14 | Virulence factor | VFGQTDETTIPLLANALADSM*NQSELETLPR | Streptococcus macedonicus, Streptococcus equinus | |
| ST3 | Virulence-associated protein E | M*KATVDNYVLVLRNDPYISESLK | Streptococcus pasteurianus | |
| ST9 | Equibactin | LYEISLKVADC*LGKNGVK | Streptococcus equi | |
| Antimicrobial production | ST3 | Bacteriocin | WTSKSSKAYAYAGQTSYAFIK | Streptococcus salivarius |
| ST2 | Bacteriocin | M*SQKIGIM*M*NIK | Streptococcus intermedius | |
| ST14 | Bacteriocin-associated integral membrane protein | AIAVGFSLAGVLAILM*QK | Streptococcus pneumoniae | |
| ST4 | LanT protein | QNVDKLHFTRFDK | Streptococcus pneumoniae | |
| ST12 | LanM protein | RAATKFM*INTDC*PSK | Streptococcus pneumoniae | |
| ABC Transporters | ST2 | Metal ABC transporter | DGADYISVM*QDNLKALEK | Streptococcus varani |
| ST6 | Metal ABC transporter | VPSAYIWEINTEEEGTPDQISSLIEK | Streptococcus pyogenes, Streptococcus equi subsp. zooepidemicus Sz105, Streptococcus canis, Streptococcus castoreus, Streptococcus porcinus, Streptococcus ictaluri, Streptococcus equi | |
| ST10 | Copper ABC transporter | SM*PDAIYLFTLLKVAC*M*GLTSFYSLR | Streptococcus infantarius, Streptococcus lutetiensis, Streptococcus equinus, Streptococcus sp. CNU 77-61, Streptococcus sp. KCJ4932 | |
| ST1 | Copper ABC transporter | NNLTLYENQYSLPIAFASQSIYNNVK | Streptococcus mitis | |
| ST10 | Zinc ABC transporter | AVIARM*FASDPNIFVLDEPTTGM*DAGSK | Streptococcus spp. | |
| ST3 | Zinc ABC transporter | TIYKNFM*EIGTAILM*STGLAISLIVM*SKGK | Streptococcus cristatus, Streptococcus sp. HMSC062B01, Streptococcus gordonii, | |
| ST2 | Cobalt or another cation ABC transporter | DGKLREVFQIPSYEM*TQVASK | Streptococcus pneumoniae | |
| ST3 | Cobalt ABC transporter | LSSDPVEVTQYYIEKGGPNV | Streptococcus salivarius | |
| ST2 | Cobalt ABC transporter (CbiM) | IISKDPNSKTM*LALSGAFIFILSSLK | Streptococcus australis, Streptococcus parasanguinis | |
| ST4 | FeoABC transporter (FeoB) | LM*DM*GLTHHTKIYLRK | Streptococcus gallolyticus | |
| ST9 | FeoABC transporter (FeoB) | EATGNQNISPNLTISNAQLNLEDKNK | Streptococcus dysgalactiae | |
| ST1 | Bacitracin ABC transporter (BceAB) | TVLGFGC*FVVQLVVIILVAYANGYVM*K | Streptococcus sp. HSISM1, Streptococcus parasanguinis | |
| ST14 | Bacitracin ABC transporter (BceAB) | QNIIALIQENGIKKSVLAK | Streptococcus sp. SK643, Streptococcus pseudopneumoniae | |
| ST2 | Bacitracin ABC transporter | SVEYPEKIATLLVNAGYPPK | Streptococcus sanguinis | |
| ST9 and ST12 | Bacteriocin ABC transporter | VNKGEFIAIM*GESGSGK | Streptococcus phocae | |
| ST9 | Bacteriocin ABC transporter | M*IVNFYTPNHGQITLGDYDLK | Streptococcus gallolyticus | |
| ST4 | Bacteriocin ABC transporter | KTVEDLSM*M*KGDM*TFK | Streptococcus oralis, Streptococcus sp. NPS 308, Streptococcus sp. oral taxon 071 str. 73H25AP, Streptococcus mitis, Streptococcus sp. VT 162, Streptococcus australis, Streptococcus pseudopneumoniae, Streptococcus halitosis, Streptococcus spp. | |
| ST9 | Lantibiotic Mutacin ABC transporter protein (MutE) | LM*VPILNILPNGLPAGTDAVVAPK | Streptococcus sobrinus | |
| ST4 | Lantibiotic ABC transporter | STIM*KIIFGLENADSGAIVFNGGKNAGK | Streptococcus mitis | |
| ST14 | Amino acid ABC transporter | M*VDGKNQVVGADIGM*AQAIADELGVK | Streptococcus oralis | |
| ST3 | Amino acid ABC transporter | NLTDKSQM*NIGIFFAIIALVVIWFLM*KK | Streptococcus parasanguinis | |
| ST13 | Amino acid ABC transporter | TGVPLLTPSGTQDDLTVDAK | Streptococcus sp. 449_SSPC, Streptococcus salivarius | |
| ST14 | Amino acid ABC transporter | VIFM*DKGIIAEEGKPEDLFTNPKEER | Streptococcus sp. oral taxon 058, Streptococcus oralis | |
| ST13 | Amino acid ABC transporter | IVLPQAFRIALPNLTTALLNLM*R | Streptococcus sp. AS14, Streptococcus sanguinis, Streptococcus cristatus, Streptococcus sp. CCH8-C6 | |
| ST9, ST13 and ST14 | Amino acid ABC transporter | NLLLAPVKVQKR | Streptococcus sp. 45, Streptococcus infantarius, Streptococcus sp. KCJ4932 Streptococcus infantarius subsp. infantarius CJ18, Streptococcus lutetiensis 033, Streptococcus infantarius, Streptococcus equinus | |
| ST10 | Glutamine ABC transporter | DASLAPM*FVAGAIYLIM*IGLVTLISKQVEK | Streptococcus sp. DD13 | |
| ST13 | Glutamine ABC transporter | KDEVIKEAENLLER | Streptococcus sanguinis | |
| ST14 | Glycine/betaine ABC transporter | YDLQVLEDDKQLFPPYQGAPLM*KEDLLK | Streptococcus oralis, Streptococcus mitis | |
| ST2 | Glycine/betaine ABC transporter | QEITLAYVEWDSEVASTNVLAEVLKTK | Streptococcus infantarius | |
| ST4 | Glycine/betaine ABC transporter | AKLRTIVAAFAVM*VLGLGASYAPSM*IPSK | Streptococcus infantis | |
| ST14 | Oligopeptide ABC transporter | KNVQM*IFQDPQASLNAR | Streptococcus infantarius, Streptococcus lutetiensis | |
| ST1 | Multidrug ABC transporter | QLQQYIYESLLTTSVK | Streptococcus suis | |
| ST4 | Multidrug ABC transporter | SGSKALKQLQQYIYESLLTTSVK | Streptococcus suis | |
| ST10 | Multidrug ABC transporter | LESKEIDENSIVSK | Streptococcus pneumoniae, Streptococcus salivarius, Streptococcus sp. HMSC068F04, Streptococcus sp. FDAARGOS_192, Streptococcus sp. SR4, thermophilus, Streptococcus sp. C150, Streptococcus sp. HMSC064H09, Streptococcus sp. HMSC064H03, Streptococcus sp. HSISS2 | |
| ST2 | Multidrug ABC transporter | AQGTLADLQATFGDASASLNDIYLALTKEV | Streptococcus phocae | |
| ST1 | Multidrug ABC transporter | YLLNLDEKQINIAPHLTINHLK | Streptococcus | |
| ST1 | Multidrug ABC transporter | M*PTAFYLFFSSM*YQDTPGGPANFM*R | Streptococcus pneumoniae | |
| ST5 | Multidrug ABC transporter | TTLIM*VSQRTNSLAK | Streptococcus sp. ‘caviae’ | |
| ST7 | Multidrug ABC transporter | FPNAFYLSM*SILLVQAVLNM*R | Streptococcus pantholopis | |
| ST3 | Multidrug ABC transporter | SGVVLSLLGAM*ISFILYLVFLKANIK | Streptococcus sp. HMSC066E07, Streptococcus anginosus | |
| Multidrug ABC transporter | IAYLPQEGALFHDTVLYNLTIGREVPEDR | Streptococcus suis | ||
| ST14 | Macrolide ABC transporter (MacB) | STLM*NIIGM*LDRPTSGEYYLEGEEVAKLSEK | Streptococcus anginosus, Streptococcus sp. KCOM 2412, Streptococcus sp. HMSC057E02 | |
| Other Transporters | ST2 | Manganese transport protein MntH | YLLLSVVLISSLIAM*QLQQM*AGKLGIVTQK | Streptococcus equinus, Streptococcus sp. KCJ4950 |
| ST6 | HlyC/CorC family transporter | TAPVIIFLGKIVSPFVWLLSASTNLLSQM*TPM*K | Streptococcus cristatus, Streptococcus sp. marseille-P644, Streptococcus sp. marseille-P7375 | |
| ST9 | Multidrug transporter MatE | AM*LIM*SLGAGINIVLDPVLM*IM*FK | Streptococcus intermedius, Streptococcus sp. AS20 | |
| ST14 | MFS Lantibiotic transporter | DLWC*NM*IIAAK | Streptococcus dysgalactiae |
| Sample | Species | Strain | Source | NCBI Accession No. of 16S RNA Gene | Total Peptides |
|---|---|---|---|---|---|
| ST1 | Streptococcus uberis | CECT 994 | unknown | JN630842.1 | 282 |
| ST2 | Streptococcus agalactiae | CECT 183 | Milk | KC510212.1 | 409 |
| ST3 | Streptococcus dysgalactiae subsp. equisimilis | CECT 926 | unknown | 259 | |
| ST4 | Streptococcus parauberis | DSM 6631 | Mastitis sample milk | NR_043001.1 | 221 |
| ST5 | Streptococcus parauberis | DSM 6632 | Raw milk | JN630844.1 | 134 |
| ST6 | Streptococcus agalactiae | USC1-LHICA | Mastitis sample milk | KP001323.1 | 140 |
| ST7 | Streptococcus agalactiae | USC3-LHICA | Mastitis sample milk | KC510215.1 | 169 |
| ST8 | Streptococcus uberis | USC5-LHICA | Mastitis sample milk | KC510216.1 | 131 |
| ST9 | Streptococcus dysgalactiae subsp. dysgalactiae | USC13-LHICA | Mastitis sample milk | KC510218.1 | 249 |
| ST10 | Streptococcus canis | USC52-LHICA | Mastitis sample milk | KC510222.1 | 247 |
| ST11 | Streptococcus uberis | USC69-LHICA | Mastitis sample milk | KC510224.1 | 44 |
| ST12 | Streptococcus gallolyticus subsp. gallolyticus | USC83-LHICA | Mastitis sample milk | KC510227.1 | 154 |
| ST13 | Streptococcus gallolyticus subsp. gallolyticus | USC84 | Mastitis sample milk | KC51022.8 | 148 |
| ST14 | Streptococcus dysgalactiae subsp. dysgalactiae | CECT 758 | Mastitis sample milk | KC51021.3 | 238 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abril, A.G.; Carrera, M.; Böhme, K.; Barros-Velázquez, J.; Rama, J.-L.R.; Calo-Mata, P.; Sánchez-Pérez, A.; Villa, T.G. Proteomic Characterization of Antibiotic Resistance, and Production of Antimicrobial and Virulence Factors in Streptococcus Species Associated with Bovine Mastitis. Could Enzybiotics Represent Novel Therapeutic Agents Against These Pathogens? Antibiotics 2020, 9, 302. https://0-doi-org.brum.beds.ac.uk/10.3390/antibiotics9060302
Abril AG, Carrera M, Böhme K, Barros-Velázquez J, Rama J-LR, Calo-Mata P, Sánchez-Pérez A, Villa TG. Proteomic Characterization of Antibiotic Resistance, and Production of Antimicrobial and Virulence Factors in Streptococcus Species Associated with Bovine Mastitis. Could Enzybiotics Represent Novel Therapeutic Agents Against These Pathogens? Antibiotics. 2020; 9(6):302. https://0-doi-org.brum.beds.ac.uk/10.3390/antibiotics9060302
Chicago/Turabian StyleAbril, Ana G., Mónica Carrera, Karola Böhme, Jorge Barros-Velázquez, José-Luis R. Rama, Pilar Calo-Mata, Angeles Sánchez-Pérez, and Tomás G. Villa. 2020. "Proteomic Characterization of Antibiotic Resistance, and Production of Antimicrobial and Virulence Factors in Streptococcus Species Associated with Bovine Mastitis. Could Enzybiotics Represent Novel Therapeutic Agents Against These Pathogens?" Antibiotics 9, no. 6: 302. https://0-doi-org.brum.beds.ac.uk/10.3390/antibiotics9060302