1. Introduction
The ability of an organism to efficiently switch between the oxidation of different energy substrates according to environmental circumstances is known as metabolic flexibility. Healthy metabolism is characterized by physiological shifts between glucose and fat oxidation in response to nutrient availability. This process maintains homeostasis in response to changing energy demands, for example, during exercise. This transition is driven by insulin activity and regulated by a cross-talk between metabolic and signaling pathways across different tissues [
1]. Skeletal muscle, as the largest contributor to insulin-mediated glucose uptake from plasma and as a major determinant of energy expenditure in resting and non-resting conditions [
2], is one of the major drivers of metabolic flexibility.
Energy metabolism is heavily involved in the aging process, not only because mitochondrial dysfunction and impaired nutrient sensing are among the main drivers of the aging process [
3], but also because all the recognized hallmarks of aging are connected to undesirable metabolic alterations [
4]. Metabolic flexibility is recognized as a feature of healthy metabolism and has been associated with longevity and a longer health span. It has also been associated with increased insulin sensitivity [
5] and lower incidence of age-related diseases, such as type 2 diabetes [
6] and cardiovascular diseases [
7]. Treatments targeting metabolic flexibility may delay the onset of aging and related comorbidities. Currently, regular physical activity and a balanced diet are still the best available treatments to increase metabolic health and to maximize health span [
8,
9].
Computational models are key to investigate the complexity of the interactions between nutrition and physical activity. Constraint-based metabolic models have been successfully used to simulate metabolic flexibility in silico by computing the Respiratory Quotient (RQ) in different nutritional conditions, for example, after a meal, during the transition between the fast and the fed state [
10,
11]. The RQ value concerns which macro nutrients are metabolized and which pathway is used for energy production. It is defined as the ratio of carbon dioxide produced by the body to oxygen consumed by the body, and it varies between values of 0.7 for pure fat metabolism and 1.0 for pure carbohydrate metabolism.
While the influence of nutrition and diet composition on metabolic flexibility is well documented [
12,
13], fewer studies have examined the role of physical activity and sedentary behaviors on metabolic flexibility. Previous studies which modeled RQ during the fast to fed transition using constraint-based models did not take the effect of energy expenditure on RQ into consideration [
11]. In this study, we propose a new description of the fast to fed transition that allows us to simulate the effect of various levels of physical activity on fuel choice in constraint-based models.
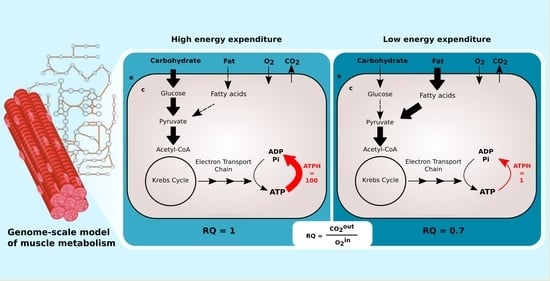
Constraint-based metabolic models do not include any description of signaling pathways. To simulate the changing concentration of plasma glucose and fatty acids after a meal, Nogiec and coworkers [
11] directly modulated the fluxes through glucose and palmitate transporters, reactions transporting substrates between external medium and cytoplasm compartments. The maximization of ATP, creatine phosphate, glycogen, and triglycerides production was used as the objective function.
This implementation is not applicable to large genome-scale metabolic models such as Recon2.2 and Recon3D, which have multiple alternative transporters for glucose and palmitate coupled with the symport or antiport of different ions, such as H
and Na
. In our model, we avoid this bias by limiting the availability of glucose and palmitate through exchange reactions to simulate the fast to fed transition. This is comparable to controlling the maximal amount of nutrients present in the external medium of a cell culture. ATP phosphodiester bond hydrolysis (ATPH) was chosen as the objective function. By maximizing ATP consumption instead of ATP production, we let the models generate ATP using the optimal pathway, thus eliminating another potential source of bias. By constraining the flux through the ATPH reaction, we can simulate a condition of reduced energy expenditure. A simplified visualization of the two models is presented in
Figure 1.
In this study, we investigate the link between physical inactivity and metabolic flexibility by simulating the effect of changing levels of energy expenditure on fuel choice, measured as RQ. Our new description of metabolic flexibility is tested in a set of constraint-based metabolic models. This model set includes two human metabolic reconstructions, Recon2.2 [
14] and Recon3D [
15], a model of central carbon metabolism, MitoCore [
16] and a set of 24 patient-derived models of skeletal muscle metabolism [
17]. We show that, in all models tested, fuel choice is sensitive to ATP consumption rate, and that a reduction in ATP consumption reproduces phenotypes associated with metabolic inflexibility.
3. Discussion
Metabolic flexibility is an important integrative biology concept which can help us understand the link between sedentary behavior, overnutrition and the dysregulation of energy metabolism and is an important part of metabolic health. Knowledge of the determinants of metabolic flexibility will help develop treatments to maintain and restore metabolic health in pathologies associated with metabolic inflexibility, such as insulin resistance, Type 2 diabetes, cardiovascular diseases, and aging.
Previous computational models of metabolic flexibility focused on nutritional intake as a determinant of metabolic flexibility, while the effect of energy expenditure on fuel choice remained understudied. In this study, we propose a new description of metabolic flexibility in genome-scale metabolic models, which enables the study of the interactions between physical activity and nutrition.
Patterns in fuel oxidation are determined not only by dietary intake but also by energy expenditure. We constrained the flux through the ATPH to reproduce a condition of lower energy expenditure. Limiting the flux through this reaction had a large effect on RQ and was sufficient to reproduce phenotypes associated with metabolic inflexibility, such as a lower RQ between the fast and fed states.
When the flux through the ATP phosphodiester bond hydrolysis (ATPH) reaction was progressively reduced, RQ values progressively increased, while the difference between RQ in the fast and fed condition decreased. Since low energy expenditure is one of the main determinants of metabolic inflexibility, a physical activity intervention should restore metabolic flexibility even in absence of a nutritional intervention. To verify this hypothesis, we simulated the fast to fed transition in a set of patient-derived models of skeletal muscle metabolism that describe the metabolism of 12 older individuals before and after a 12-week resistance training program [
24]. In high energy expenditure conditions, all models had the same response during the fast to fed transition. In low energy expenditure conditions (ATPH upper bound = 35 mM/gDw/h), trained models showed an increased utilization of the oxidative phosphorylation (OXPHOS) pathway for energy production (
Figure 6). These results show that patient-derived models can capture some of the long-term metabolic adaptations resulting from a metabolic intervention, supporting the idea that these models can be used to improve our understanding of individual responses to diet and exercise.
Constraint-based metabolic models are useful tools to investigate the interactions between physical activity and nutrition, and how they influence metabolic health and the aging process. Nevertheless, there are still important inconsistencies in the predictions of different models. FBA predictions are heavily biased by the reaction composition of the models and by the boundary constraints the modelers apply. This fact is often overlooked in genome-scale metabolic modeling studies. Unfortunately, this is necessary due to the characteristics of the genome-scale modeling framework itself, especially when dealing with larger human models such as Recon2.2 and Recon3D, which contain a large number of boundary reactions. Leaving these models under-constrained often causes the appearance of physically unfeasible and physiologically unrealistic flux cycles in the FBA solution. We applied constraints to boundary reactions and used methods such as parsimonious FBA (PFBA) to reduce this effect. The differences that can be observed in the results are also due to the fact that different models have different focuses: MitoCore is limited to central carbon metabolism, Recon models are “generic” metabolic reconstructions, and skeletal muscle models collectively represent a tissue-specific metabolism. It is important to remember that none of these models are “true”; they are different approximations of human physiology.
However, we can identify properties that are common to all models: RQ predictions are dependent on the upper bound of ATPH reactions, even when palmitate and glucose intake fluxes are kept constant. Moreover, RQ change is independent of endocrine (insulin) signaling and allosteric regulation, which cannot be described in GSMMs. Metabolic flexibility thus appears to be an intrinsic property of the metabolic network, which is determined not only by intake fluxes but also by energy expenditure and by the model-specific ATP yields of relevant substrates.
In principle, the calculation of RQ from reaction stoichiometry is straightforward. In practice, it is difficult to obtain reproducible results in RQ simulations across different metabolic models.
These simulations are challenging not only because of their sensitivity to ‘technical’ variability, for example, the use of a different solver software, or due to different model composition and constraints on intake fluxes, but also because they are sensitive to "biological" variability, for example, different nutrition and energy expenditure habits, or different genetic backgrounds among different individuals. It may be difficult to distinguish "technical" variability from "biological" variability. Several model inconsistencies that biased the results were addressed, as discussed in
Appendix A. To ensure the reproducibility of the results, model composition and simulation parameters such as reaction bounds should be standardized as much as possible.
Metabolic flexibility is an important health concept that integrates nutrition and energy expenditure. Expanding this concept to any response of fuel metabolism to external stressors, such as hot and cold temperatures, traumatic events such as illness, injuries or surgeries, and psychological stress [
25] could reveal more details about how these factors interact in many pathological and physiological conditions, including aging.
Author Contributions
Conceptualization, A.C. and N.A.W.v.R.; methodology, A.C.; original draft preparation, A.C.; review and editing, N.A.W.v.R.; supervision, P.A.J.H. and N.A.W.v.R.; funding acquisition, N.A.W.v.R. All authors have read and agreed to the published version of the manuscript.
Funding
This project has received funding from the European Union’s Horizon 2020 research and innovation program, under the Marie Sklodowska-Curie grant agreement 675003.
Institutional Review Board Statement
Since it is a retrospective analysis of publicly available data, ethical review and approval were waived for this study.
Informed Consent Statement
Since this study involves the use of anonymized data downloaded from a public repository (Gene Expression Omnibus, GEO), patient consent was waived.
Data Availability Statement
The gene expression data presented in this study are openly available in GEO, the Gene Expression Omnibus at accession number GSE28422.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| RQ | Respiratory Quotient |
| ATPH | ATP phosphodiester bond hydrolysis reaction |
| OXPHOS | Oxidative phosphorylation |
| UB | Upper bound |
| gDw | Grams of dry weight |
References
- Smith, R.L.; Soeters, M.R.; Wüst, R.C.; Houtkooper, R.H. Metabolic flexibility as an adaptation to energy resources and requirements in health and disease. Endocr. Rev. 2018, 39, 489–517. [Google Scholar] [CrossRef] [Green Version]
- Zurlo, F.; Larson, K.; Bogardus, C.; Ravussin, E. Skeletal muscle metabolism is a major determinant of resting energy expenditure. J. Clin. Investig. 1990, 86, 1423–1427. [Google Scholar] [CrossRef] [Green Version]
- López-Otín, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The hallmarks of aging. Cell 2013, 153, 1194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- López-Otín, C.; Galluzzi, L.; Freije, J.M.; Madeo, F.; Kroemer, G. Metabolic Control of Longevity. Cell 2016, 166, 802–821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galgani, J.E.; Moro, C.; Ravussin, E. Metabolic flexibility and insulin resistance. Am. J. Physiol.-Endocrinol. Metab. 2008, 295, 1009–1017. [Google Scholar] [CrossRef] [Green Version]
- Corpeleijn, E.; Saris, W.H.; Blaak, E.E. Metabolic flexibility in the development of insulin resistance and type 2 diabetes: Effects of lifestyle: Etiology and Pathophysiology. Obes. Rev. 2009, 10, 178–193. [Google Scholar] [CrossRef]
- Vallerie, S.N.; Bornfeldt, K.E. Metabolic flexibility and dysfunction in cardiovascular cells. Arterioscler. Thromb. Vasc. Biol. 2015, 35, e37–e42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Whittaker, A.C.; Delledonne, M.; Finni, T.; Garagnani, P.; Greig, C.; Kallen, V.; Kokko, K.; Lord, J.; Maier, A.B.; Meskers, C.G.; et al. Physical Activity and Nutrition INfluences In ageing (PANINI): Consortium mission statement. Aging Clin. Exp. Res. 2018, 30, 685–692. [Google Scholar] [CrossRef] [PubMed]
- Mercken, E.M.; Carboneau, B.A.; Krzysik-Walker, S.M.; De Cabo, R. Of mice and men: The benefits of caloric restriction, exercise, and mimetics. Ageing Res. Rev. 2012, 11, 390–398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Väremo, L.; Scheele, C.; Broholm, C.; Mardinoglu, A.; Kampf, C.; Asplund, A.; Nookaew, I.; Uhlén, M.; Pedersen, B.K.; Nielsen, J. Proteome- and Transcriptome-Driven Reconstruction of the Human Myocyte Metabolic Network and Its Use for Identification of Markers for Diabetes. Cell Rep. 2015, 11, 921–933. [Google Scholar] [CrossRef] [Green Version]
- Nogiec, C.; Burkart, A.; Dreyfuss, J.M.; Lerin, C.; Kasif, S.; Patti, M.E. Metabolic modeling of muscle metabolism identifies key reactions linked to insulin resistance phenotypes. Mol. Metab. 2015, 4, 151–163. [Google Scholar] [CrossRef]
- Kelley, D.E.; Mandarino, L.J. Fuel selection in human skeletal muscle in insulin resistance: A reexamination. Diabetes 2000, 49, 677–683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Herpen, N.A.; Schrauwen-Hinderling, V.B.; Schaart, G.; Mensink, R.P.; Schrauwen, P. Three weeks on a high-fat diet increases intrahepatic lipid accumulation and decreases metabolic flexibility in healthy overweight men. J. Clin. Endocrinol. Metab. 2011, 96, 691–695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swainston, N.; Smallbone, K.; Hefzi, H.; Dobson, P.D.; Brewer, J.; Hanscho, M.; Zielinski, D.C.; Ang, K.S.; Gardiner, N.J.; Gutierrez, J.M.; et al. Recon2.2: From reconstruction to model of human metabolism. Metabolomics 2016, 12. [Google Scholar] [CrossRef]
- Brunk, E.; Sahoo, S.; Zielinski, D.C.; Altunkaya, A.; Dräger, A.; Mih, N.; Gatto, F.; Nilsson, A.; Preciat Gonzalez, G.A.; Aurich, M.K.; et al. Recon3D enables a three-dimensional view of gene variation in human metabolism. Nat. Biotechnol. 2018, 36, 272–281. [Google Scholar] [CrossRef]
- Smith, A.C.; Eyassu, F.; Mazat, J.P.; Robinson, A.J. MitoCore: A curated constraint-based model for simulating human central metabolism. BMC Syst. Biol. 2017, 11, 114. [Google Scholar] [CrossRef] [Green Version]
- Cabbia, A.; Hilbers, P.A.; van Riel, N.A. A Distance-Based Framework for the Characterization of Metabolic Heterogeneity in Large Sets of Genome-Scale Metabolic Models. Patterns 2020, 1, 100080. [Google Scholar] [CrossRef]
- Lewis, N.E.; Hixson, K.K.; Conrad, T.M.; Lerman, J.A.; Charusanti, P.; Polpitiya, A.D.; Adkins, J.N.; Schramm, G.; Purvine, S.O.; Lopez-Ferrer, D.; et al. Omic data from evolved E. coli are consistent with computed optimal growth from genome-scale models. Mol. Syst. Biol. 2010, 6. [Google Scholar] [CrossRef]
- Tareen, S.H.; Kutmon, M.; Adriaens, M.E.; Mariman, E.C.; de Kok, T.M.; Arts, I.C.; Evelo, C.T. Exploring the cellular network of metabolic flexibility in the adipose tissue. Genes Nutr. 2018, 13, 17. [Google Scholar] [CrossRef]
- Gawron, P.; Ostaszewski, M.; Satagopam, V.; Gebel, S.; Mazein, A.; Kuzma, M.; Zorzan, S.; McGee, F.; Otjacques, B.; Balling, R.; et al. MINERVA—A platform for visualization and curation of molecular interaction networks. NPJ Syst. Biol. Appl. 2016, 2, 16020. [Google Scholar] [CrossRef] [Green Version]
- Sivitz, W.I.; Yorek, M.A. Mitochondrial dysfunction in diabetes: From molecular mechanisms to functional significance and therapeutic opportunities. Antioxid. Redox Signal. 2010, 12, 537–577. [Google Scholar] [CrossRef] [Green Version]
- Toledo, F.G.; Menshikova, E.V.; Ritov, V.B.; Azuma, K.; Radikova, Z.; DeLany, J.; Kelley, D.E. Effects of physical activity and weight loss on skeletal muscle mitochondria and relationship with glucose control in type 2 diabetes. Diabetes 2007, 56, 2142–2147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bordenave, S.; Metz, L.; Flavier, S.; Lambert, K.; Ghanassia, E.; Dupuy, A.M.; Michel, F.; Puech-Cathala, A.M.; Raynaud, E.; Brun, J.F.; et al. Training-induced improvement in lipid oxidation in type 2 diabetes mellitus is related to alterations in muscle mitochondrial activity. Effect of endurance training in type 2 diabetes. Diabetes Metab. 2008, 34, 162–168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raue, U.; Trappe, T.A.; Estrem, S.T.; Qian, H.R.; Helvering, L.M.; Smith, R.C.; Trappe, S. Transcriptome signature of resistance exercise adaptations: Mixed muscle and fiber type specific profiles in young and old adults. J. Appl. Physiol. 2012, 112, 1625–1636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rynders, C.A.; Blanc, S.; DeJong, N.; Bessesen, D.H.; Bergouignan, A. Sedentary behaviour is a key determinant of metabolic inflexibility. J. Physiol. 2018, 596, 1319–1330. [Google Scholar] [CrossRef]
- Schultz, A.; Qutub, A.A. Reconstruction of Tissue-Specific Metabolic Networks Using CORDA. PLoS Comput. Biol. 2016, 12, 1–33. [Google Scholar] [CrossRef] [PubMed]
- Wegrzyn, A.B.; Stolle, S.; Rienksma, R.A.; Martins dos Santos, V.A.; Bakker, B.M.; Suarez-Diez, M. Cofactors revisited—Predicting the impact of flavoprotein-related diseases on a genome scale. Biochim. Biophys. Acta Mol. Basis Dis. 2019, 1865, 360–370. [Google Scholar] [CrossRef]
- Ebrahim, A.; Lerman, J.A.; Palsson, B.O.; Hyduke, D.R. COBRApy: COnstraints-Based Reconstruction and Analysis for Python. BMC Syst. Biol. 2013, 7, 1. [Google Scholar] [CrossRef] [Green Version]
| Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).