Digital Soil Mapping over Large Areas with Invalid Environmental Covariate Data
Abstract
:1. Introduction
2. Methods
2.1. Basic Idea
2.2. Detailed Design of the Proposed Method
3. Case Study
3.1. Study Area and Data
3.2. Experimental Design
3.3. Evaluation Method
3.4. Results and Discussion
3.4.1. Under the Cell-Level Test Scenarios
3.4.2. Under the Block-Level Test Scenarios
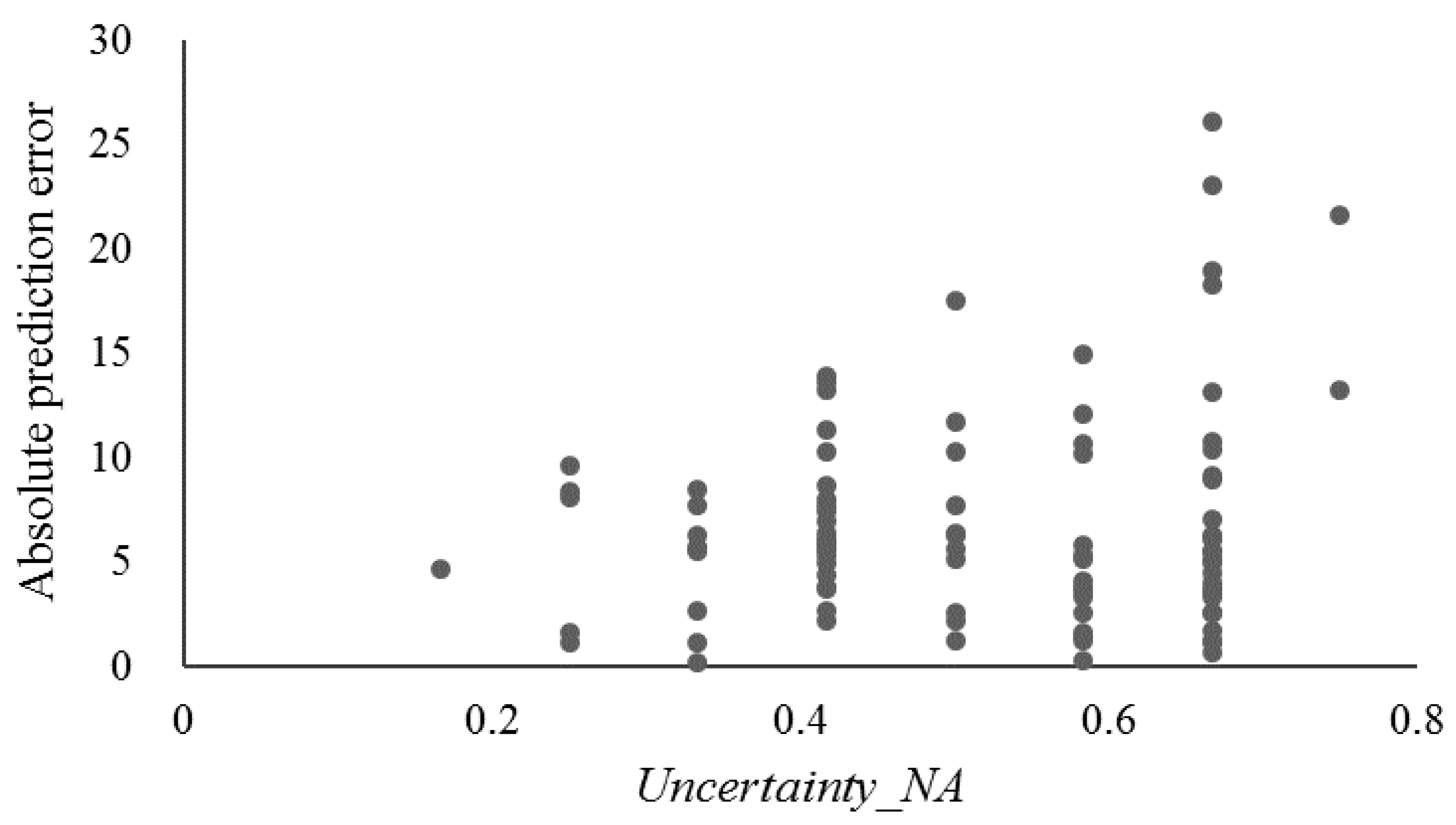
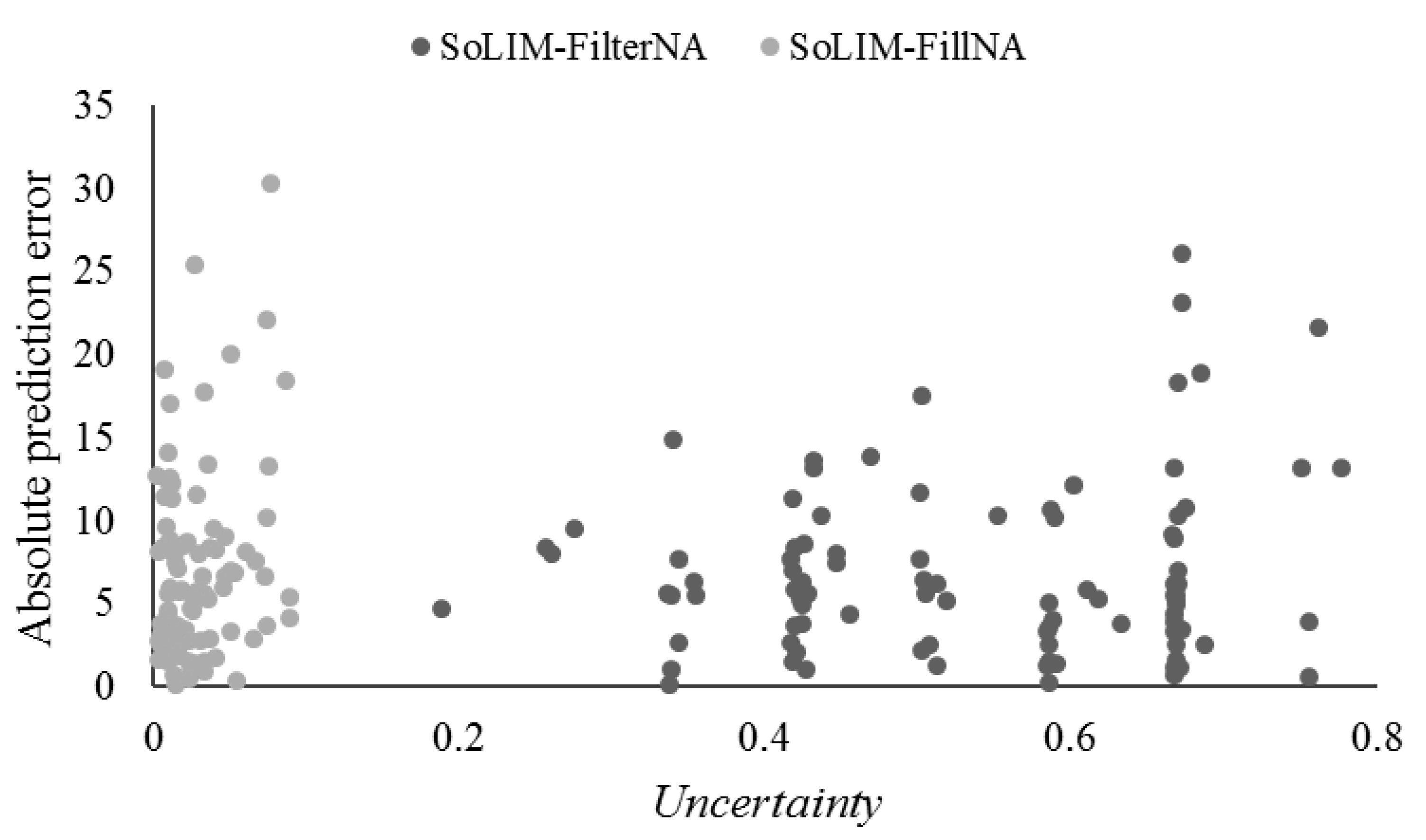
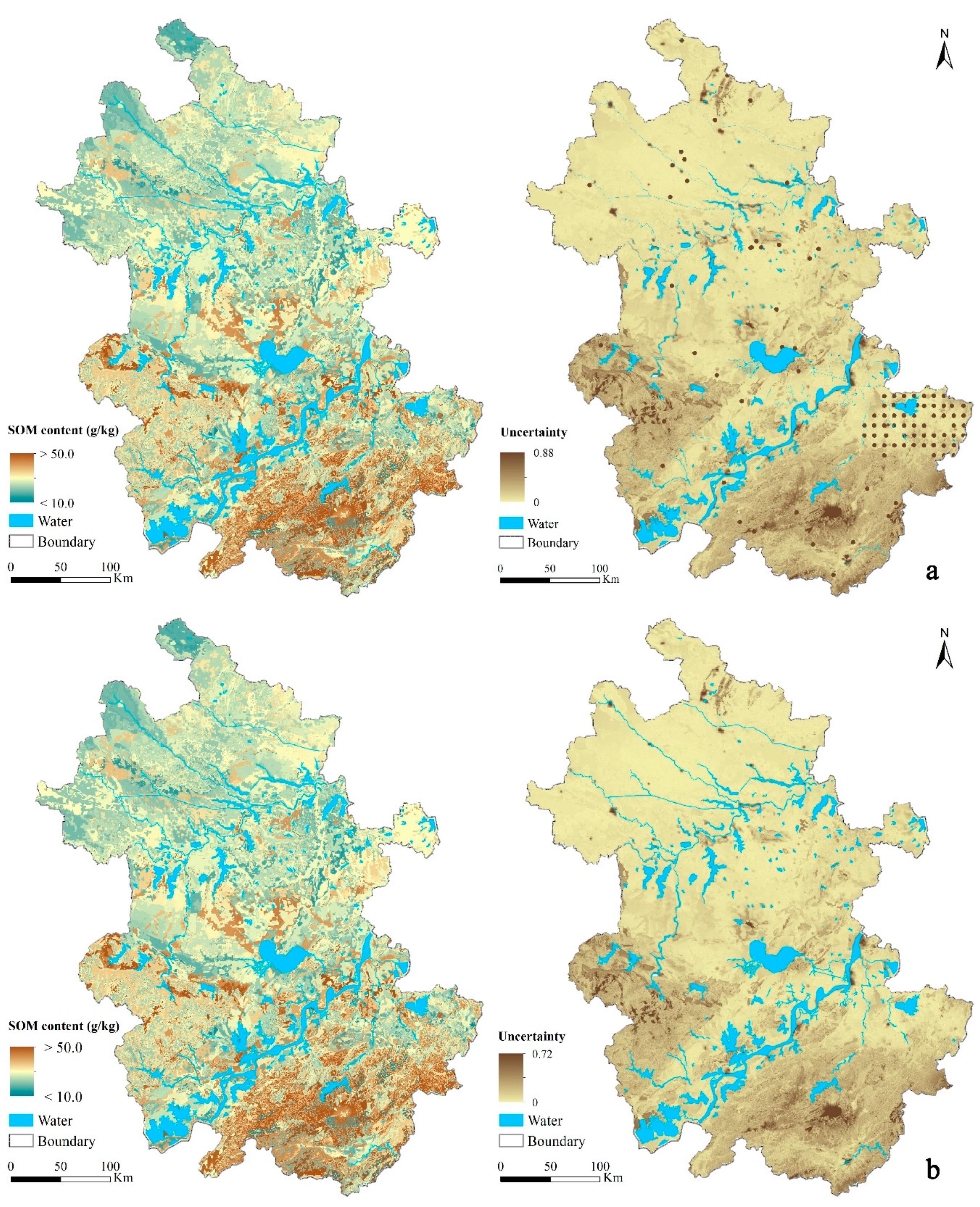
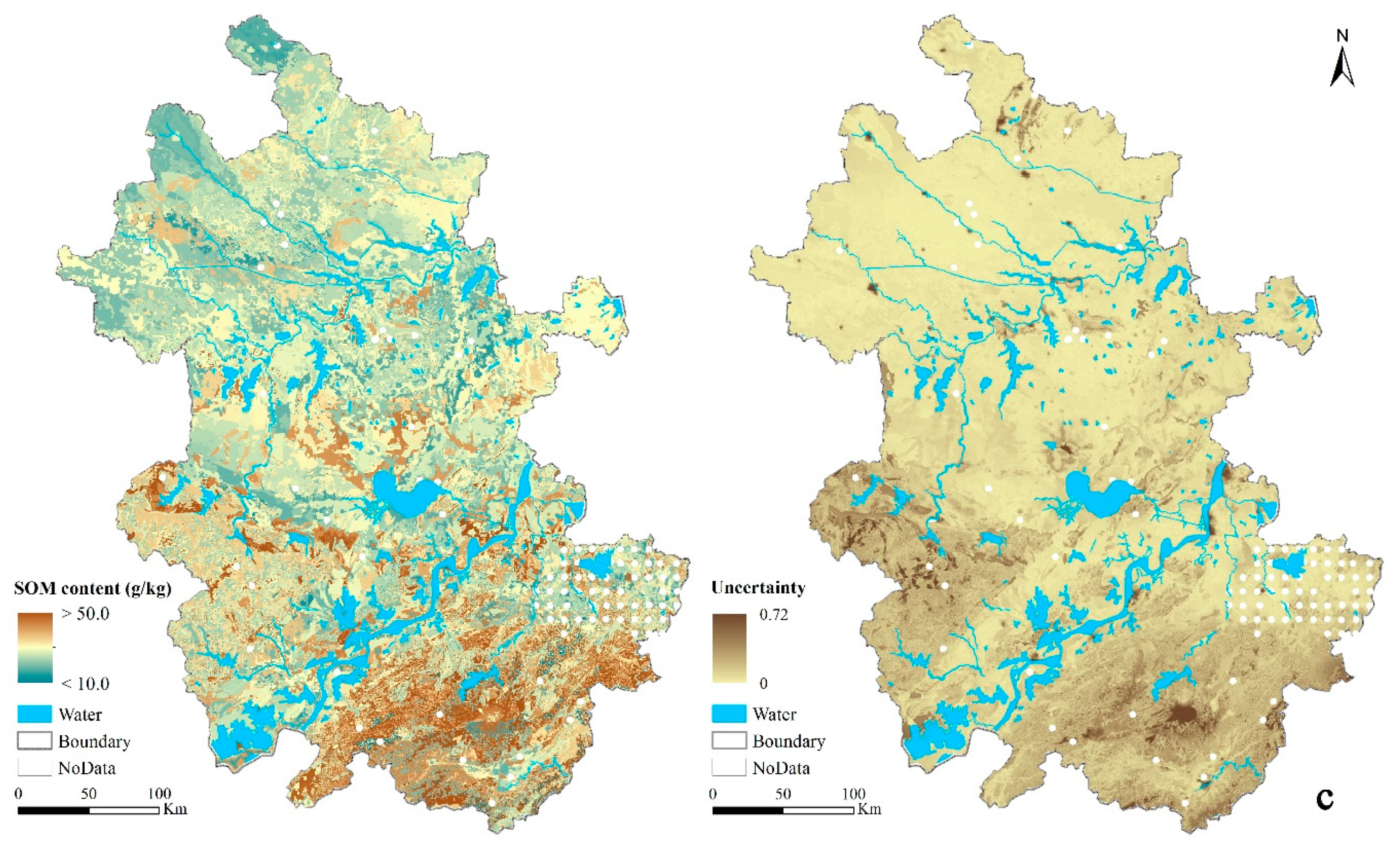
3.4.3. Prediction Uncertainty
4. Conclusions and Future Work
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Goodchild, M.F.; Parks, B.O.; Steyaert, L.T. Environmental Modeling with GIS; Oxford University Press: New York, NY, USA, 1993. [Google Scholar]
- Shani, U.; Ben-Gal, A.; Tripler, E.; Dudley, L.M. Plant response to the soil environment: An analytical model integrating yield, water, soil type, and salinity. Water Resour. Res. 2007, 43, W08418. [Google Scholar] [CrossRef]
- Grunwald, S.; Thompson, J.; Boettinger, J. Digital soil mapping and modeling at continental scales: Finding solutions for global issues. Soil Sci. Soc. Am. J. 2011, 75, 1201–1213. [Google Scholar] [CrossRef]
- Stoorvogel, J.J.; Bakkenes, M.; Temme, A.J.; Batjes, N.H.; ten Brink, B.J. S-world: A global soil map for environmental modelling. Land Degrad. Dev. 2017, 28, 22–33. [Google Scholar] [CrossRef]
- McBratney, A.B.; Santos, M.L.M.; Minasny, B. On digital soil mapping. Geoderma 2003, 117, 3–52. [Google Scholar] [CrossRef]
- Zhu, A.X.; Hudson, B.; Burt, J.; Lubich, K.; Simonson, D. Soil mapping using GIS, expert knowledge, and fuzzy logic. Soil Sci Soc. Am. J. 2001, 65, 1463–1472. [Google Scholar] [CrossRef] [Green Version]
- Minasny, B.; McBratney, A.B. Digital soil mapping: A brief history and some lessons. Geoderma 2016, 264, 301–311. [Google Scholar] [CrossRef]
- Zhu, A.X.; Band, L.; Vertessy, R.; Dutton, B. Derivation of soil properties using a soil land inference model (SoLIM). Soil Sci Soc. Am. J. 1997, 61, 523–533. [Google Scholar] [CrossRef]
- Ishioka, T. Imputation of missing values for semi-supervised data using the proximity in random forests. In Proceedings of the 14th International Conference on Information Integration and Web-based Applications & Services, Bali, Indonesia, 3–5 December 2012; pp. 319–322. [Google Scholar]
- Taghizadeh-Mehrjardi, R.; Minasny, B.; Sarmadian, F.; Malone, B. Digital mapping of soil salinity in Ardakan region, central Iran. Geoderma 2014, 213, 15–28. [Google Scholar] [CrossRef]
- Little, R.J.; Rubin, D.B. Statistical Analysis with Missing Data, 2nd ed.; John Wiley & Sons: Hoboken, NJ, USA, 2019. [Google Scholar]
- Hugelius, G.; Tarnocai, C.; Broll, G.; Canadell, J.; Kuhry, P.; Swanson, D. The Northern Circumpolar Soil Carbon Database: Spatially distributed datasets of soil coverage and soil carbon storage in the northern permafrost regions. Earth Syst. Sci. Data 2013, 5, 3–13. [Google Scholar] [CrossRef] [Green Version]
- Hengl, T.; Gruber, S.; Shrestha, D.P. Reduction of errors in digital terrain parameters used in soil-landscape modelling. Int. J. Appl. Earth Obs. Geoinf. 2004, 5, 97–112. [Google Scholar] [CrossRef]
- Grimm, R.; Behrens, T.; Marker, M.; Elsenbeer, H. Soil organic carbon concentrations and stocks on Barro Colorado Island - Digital soil mapping using Random Forests analysis. Geoderma 2008, 146, 102–113. [Google Scholar] [CrossRef]
- Hengl, T.; Heuvelink, G.B.; Kempen, B.; Leenaars, J.G.; Walsh, M.G.; Shepherd, K.D.; Sila, A.; MacMillan, R.A.; Mendes de Jesus, J.; Tamene, L.; et al. Mapping Soil Properties of Africa at 250 m Resolution: Random Forests Significantly Improve Current Predictions. PLoS ONE 2015, 10, e0125814. [Google Scholar] [CrossRef] [PubMed]
- Vågen, T.G.; Winowiecki, L.A.; Tondoh, J.E.; Desta, L.T.; Gumbricht, T. Mapping of soil properties and land degradation risk in Africa using MODIS reflectance. Geoderma 2016, 263, 216–225. [Google Scholar] [CrossRef] [Green Version]
- McBratney, A.B.; Walvoort, D.J.J. Generalised Linear Model Kriging: A generic framework for kriging with secondary data. In Proceedings of the Pedometrics 2001 4th Conference of the Working Group on Pedometric of the IUSS, Ghent, Belgium, 19–21 September 2001. [Google Scholar]
- Hengl, T.; de Jesus, J.M.; MacMillan, R.A.; Batjes, N.H.; Heuvelink, G.B.; Ribeiro, E.; Samuel-Rosa, A.; Kempen, B.; Leenaars, J.G.; Walsh, M.G.; et al. SoilGrids1km—global soil information based on automated mapping. PLoS ONE 2014, 9, e105992. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vaysse, K.; Lagacherie, P. Evaluating digital soil mapping approaches for mapping GlobalSoilMap soil properties from legacy data in Languedoc-Roussillon (France). Geoderma Reg. 2015, 4, 20–30. [Google Scholar] [CrossRef]
- Hengl, T.; Mendes de Jesus, J.; Heuvelink, G.B.; Ruiperez Gonzalez, M.; Kilibarda, M.; Blagotic, A.; Shangguan, W.; Wright, M.N.; Geng, X.; Bauer-Marschallinger, B.; et al. SoilGrids 250 m: Global gridded soil information based on machine learning. PLoS ONE 2017, 12, e0169748. [Google Scholar] [CrossRef] [Green Version]
- Ließ, M. Sampling for regression-based digital soil mapping: Closing the gap between statistical desires and operational applicability. Spat. Stat. 2015, 13, 106–122. [Google Scholar] [CrossRef]
- Zhu, A.X.; Liu, J.; Du, F.; Zhang, S.J.; Qin, C.Z.; Burt, J.; Behrens, T.; Scholten, T. Predictive soil mapping with limited sample data. Eur. J. Soil Sci. 2015, 66, 535–547. [Google Scholar] [CrossRef]
- Qin, C.Z.; Zhu, A.X.; Qiu, W.L.; Lu, Y.J.; Li, B.L.; Pei, T. Mapping soil organic matter in small low-relief catchments using fuzzy slope position information. Geoderma 2012, 171–172, 64–74. [Google Scholar] [CrossRef]
- Zhu, A.X.; Qi, F.; Moore, A.; Burt, J.E. Prediction of soil properties using fuzzy membership values. Geoderma 2010, 158, 199–206. [Google Scholar] [CrossRef]
- Zhu, A.X.; Lü, G.N.; Liu, J.; Qin, C.Z.; Zhou, C.H. Spatial prediction based on Third Law of Geography. Ann. GIS 2018, 24, 225–240. [Google Scholar] [CrossRef]
- Yang, L.; Zhu, A.X.; Zhao, Y.G.; Li, D.C.; Zhang, G.L.; Zhang, S.J.; Band, L.E. Regional Soil Mapping Using Multi-Grade Representative Sampling and a Fuzzy Membership-Based Mapping Approach. Pedosphere 2017, 27, 344–357. [Google Scholar] [CrossRef]
- An, Y.M.; Yang, L.; Zhu, A.X.; Qin, C.Z.; Shi, J.J. Identification of representative samples from existing samples for digital soil mapping. Geoderma 2018, 311, 109–119. [Google Scholar] [CrossRef]
- Zhu, A.X.; Band, L.E. A knowledge-based approach to data integration for soil mapping. Can. J. Remote Sens. 1994, 20, 408–418. [Google Scholar] [CrossRef]
- Zhu, A.X. A personal construct-based knowledge acquisition process for natural resource mapping. Int. J. Geogr. Inf. Sci. 1999, 13, 119–141. [Google Scholar] [CrossRef]
- Minasny, B.; McBratney, A.B.; Malone, B.P.; Wheeler, I. Digital Mapping of Soil Carbon. Adv. Agron. 2013, 118, 1–47. [Google Scholar]
- Zhu, A.X. Measuring uncertainty in class assignment for natural resource maps under fuzzy logic. Photogramm. Eng. Remote Sens. 1997, 63, 1195–1202. [Google Scholar]
- Qin, C.Z.; Lu, Y.J.; Bao, L.L.; Zhu, A.X.; Qiu, W.L.; Cheng, W.M. Simple digital terrain analysis software (SimDTA 1.0) and its application in fuzzy classification of slope positions. J. Geo-Inf. Sci. 2009, 11, 737–743, (in Chinese with English abstract). [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Liaw, A.; Wiener, M. Classification and regression by randomForest. R News 2002, 2, 18–22. [Google Scholar]
- Genuer, R.; Poggi, J.-M.; Tuleau-Malot, C. Variable selection using random forests. Pattern Recognit. Lett. 2010, 31, 2225–2236. [Google Scholar] [CrossRef] [Green Version]
- Pantanowitz, A.; Marwala, T. Evaluating the Impact of Missing Data Imputation through the use of the Random Forest Algorithm. arXiv 2008, arXiv:0812.2412. [Google Scholar]
- Rodriguez-Galiano, V.F.; Ghimire, B.; Rogan, J.; Chica-Olmo, M.; Rigol-Sanchez, J.P. An assessment of the effectiveness of a random forest classifier for land-cover classification. J. Photogramm. Remote Sens. 2012, 67, 93–104. [Google Scholar] [CrossRef]
- Mentch, L.; Hooker, G. Quantifying Uncertainty in Random Forests via Confidence Intervals and Hypothesis Tests. J. Mach. Learn. Res. 2016, 17, 1–41. [Google Scholar]
- Vaysse, K.; Lagacherie, P. Using quantile regression forest to estimate uncertainty of digital soil mapping products. Geoderma 2017, 291, 55–64. [Google Scholar] [CrossRef]
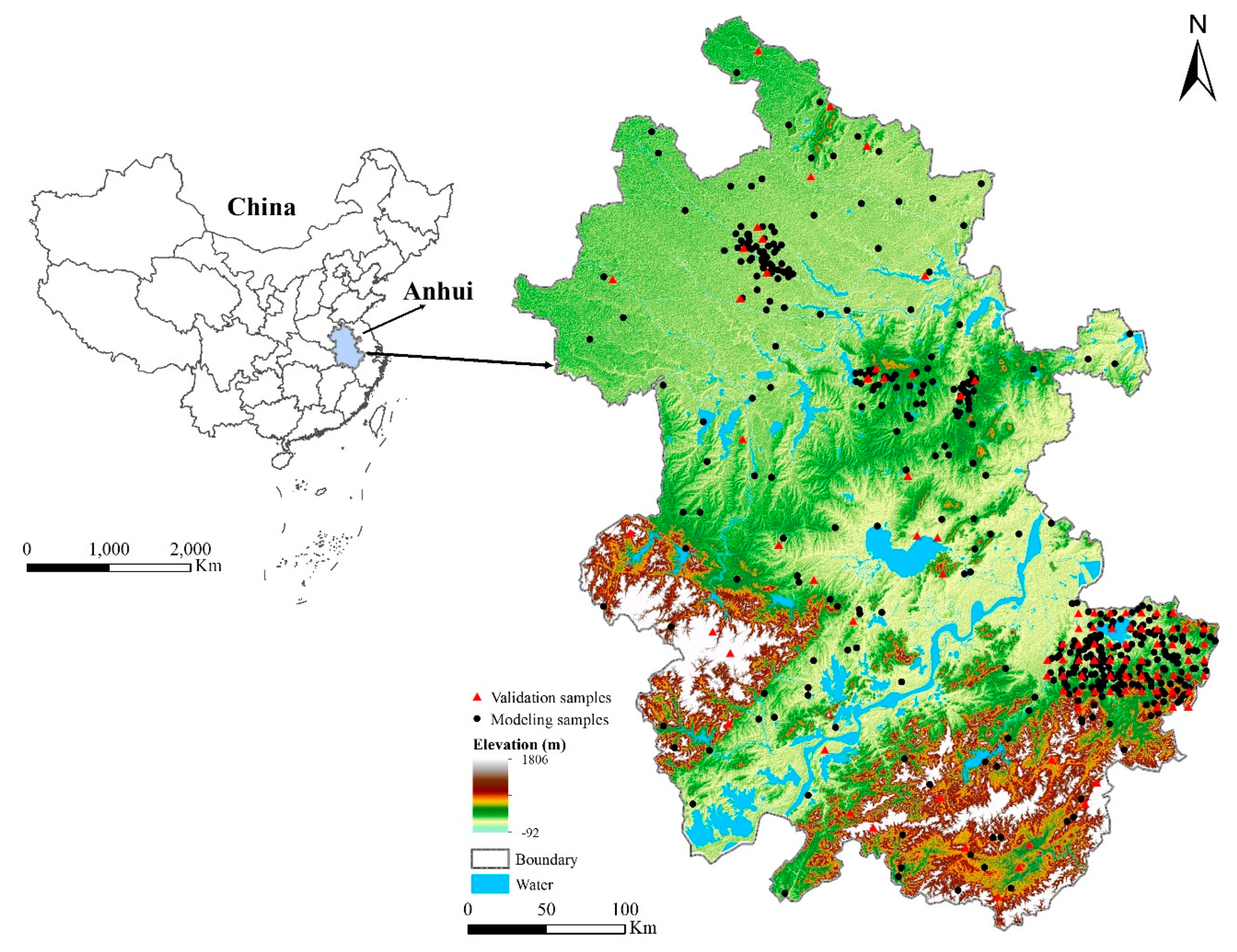
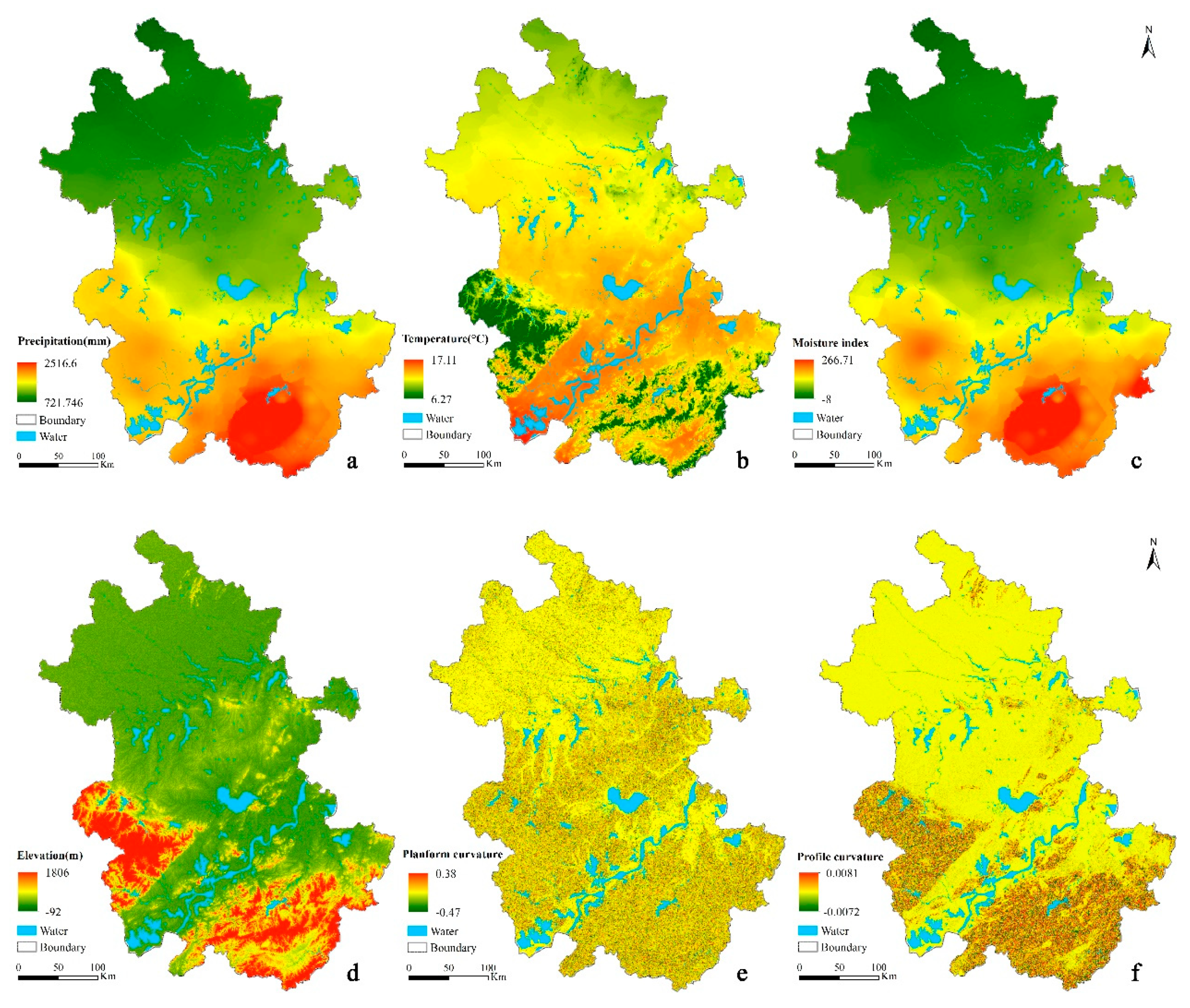
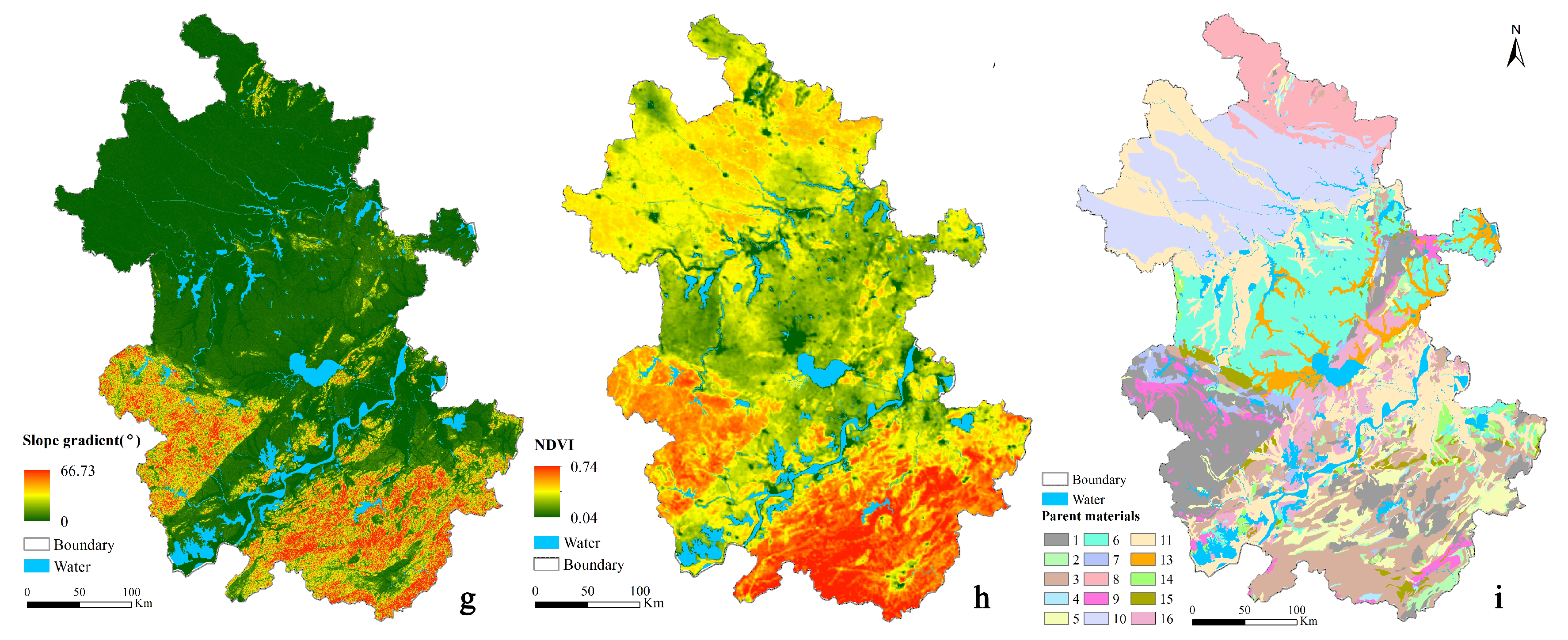
| Environmental Factor | Environmental Covariates | Data Type | Data Source | Original Resolution | Algorithm |
|---|---|---|---|---|---|
| Climate | Annual averaged precipitation | Continuous | Observations from National Meteorological station | Station | IDW |
| Annual averaged temperature | |||||
| Moisture index | Continuous | http://www.resdc.cn | 500 m | Resample | |
| Terrain | Elevation | Continuous | SRTM DEM | 90 m | -- |
| Slope gradient | Continuous | SRTM DEM | 90 m | SimDTA [32] | |
| Planform curvature | |||||
| Profile curvature | |||||
| Vegetation | NDVI | Continuous | MODIS | 250 m | Resample |
| Parent material | Parent material | Categorical | http://www.ngac.org.cn | 1:500,000 | Resample |
| Test Scenario | Level | Covariate Setting NoData | Count of Cells with NoData Set on at Least One Covariate | |
|---|---|---|---|---|
| Count | Date Type | |||
| T(V1C) | Cell-level | 1 | Continuous | 109 (i.e., all independent evaluation points) |
| T(V1T) | Cell-level | 1 | Categorical (Type) | 109 |
| T(V2) | Cell-level | 2 | Random | 109 |
| T(V3) | Cell-level | 3 | Random | 109 |
| T(V4) | Cell-level | 4 | Random | 109 |
| T(V5) | Cell-level | 5 | Random | 109 |
| T(Vr) | Cell-level | 1~5 | Random | 109 |
| T(Vr-74cell) | Cell-level | 1~5 | Random | 74 (evaluation points randomly selected) |
| T(Vr-buffer5) | Block-level | same as T(Vr) | 109 evaluation points with their buffer of 5 cells | |
| T(Vr-buffer10) | Block-level | same as T(Vr) | 109 evaluation points with their buffer of 10 cells | |
| T(Vr-buffer15) | Block-level | same as T(Vr) | 109 evaluation points with their buffer of 15 cells | |
| T(Vr-buffer25) | Block-level | same as T(Vr) | 109 evaluation points with their buffer of 25 cells | |
| Methods | Error Statistics | Test Scenario | |||||||
|---|---|---|---|---|---|---|---|---|---|
| T(V1C) | T(V1T) | T(V2) | T(V3) | T(V4) | T(V5) | T(Vr) | T(Vr-74cell) | ||
| SoLIM-FilterNA | RMSE | 8.334 | 8.253 | 8.447 | 8.654 | 8.666 | 8.681 | 8.556 | 9.052 |
| MAE | 6.786 | 6.580 | 6.727 | 6.850 | 6.916 | 6.982 | 6.877 | 7.179 | |
| SoLIM-FillNA | RMSE | 8.861 | 8.866 | 9.056 | 9.054 | 9.056 | 9.071 | 9.058 | – |
| MAE | 6.915 | 6.921 | 7.061 | 7.057 | 7.059 | 7.102 | 7.064 | – | |
| RF | RMSE | 8.414 | 8.602 | 8.682 | 8.703 | 8.710 | 8.727 | 8.660 | – |
| MAE | 6.641 | 6.897 | 7.057 | 7.027 | 6.975 | 6.733 | 7.038 | – | |
| original SoLIM | RMSE | – | – | – | – | – | – | – | 9.564 |
| MAE | – | – | – | – | – | – | – | 7.816 | |
| Methods | Error Statistics | Block-Level Test Scenario | |||
|---|---|---|---|---|---|
| T(Vr-Buffer5) | T(Vr-Buffer10) | T(Vr-Buffer15) | T(Vr-Buffer25) | ||
| SoLIM-FilterNA | RMSE | 8.556 | 8.556 | 8.556 | 8.556 |
| MAE | 6.877 | 6.877 | 6.877 | 6.877 | |
| SoLIM-FillNA | RMSE | 9.145 | 9.183 | 9.512 | 10.199 |
| MAE | 7.133 | 7.210 | 7.329 | 7.278 | |
| RF | RMSE | 9.262 | 9.325 | 9.470 | 9.655 |
| MAE | 7.532 | 7.619 | 7.793 | 8.254 | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, N.-Q.; Zhu, A.-X.; Qin, C.-Z.; Liang, P. Digital Soil Mapping over Large Areas with Invalid Environmental Covariate Data. ISPRS Int. J. Geo-Inf. 2020, 9, 102. https://0-doi-org.brum.beds.ac.uk/10.3390/ijgi9020102
Fan N-Q, Zhu A-X, Qin C-Z, Liang P. Digital Soil Mapping over Large Areas with Invalid Environmental Covariate Data. ISPRS International Journal of Geo-Information. 2020; 9(2):102. https://0-doi-org.brum.beds.ac.uk/10.3390/ijgi9020102
Chicago/Turabian StyleFan, Nai-Qing, A-Xing Zhu, Cheng-Zhi Qin, and Peng Liang. 2020. "Digital Soil Mapping over Large Areas with Invalid Environmental Covariate Data" ISPRS International Journal of Geo-Information 9, no. 2: 102. https://0-doi-org.brum.beds.ac.uk/10.3390/ijgi9020102





