CK2 Regulation: Perspectives in 2021
Abstract
:1. Introduction
1.1. Protein Kinase CK2
1.1.1. Background
1.1.2. CK2 in Disease
Okur-Chung Syndrome
COVID-19
1.1.3. CK2: The Constitutively Active Kinase
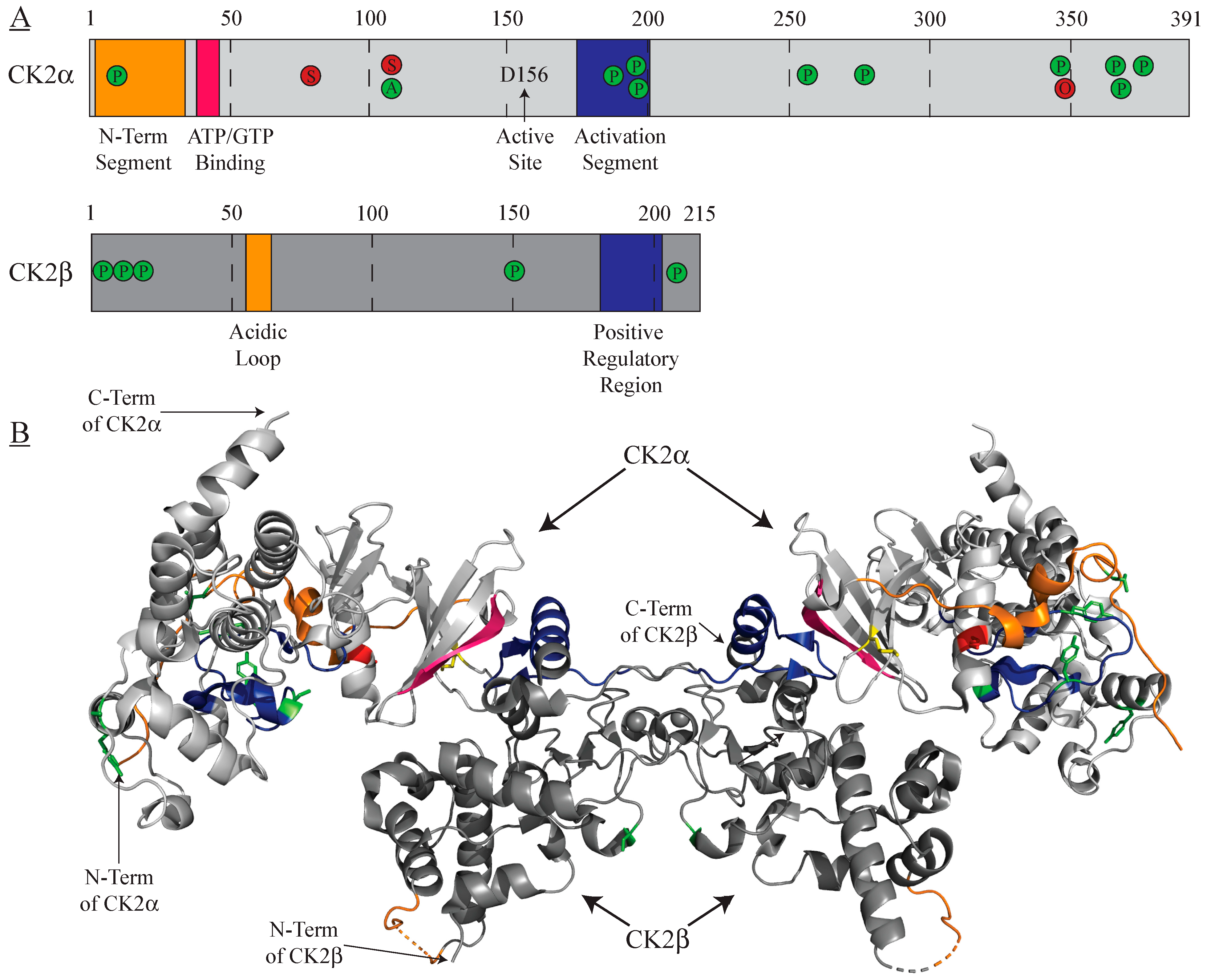
2. Intrinsic Regulation
2.1. Post-Translational Modifications
2.2. Protein–Protein Interactions
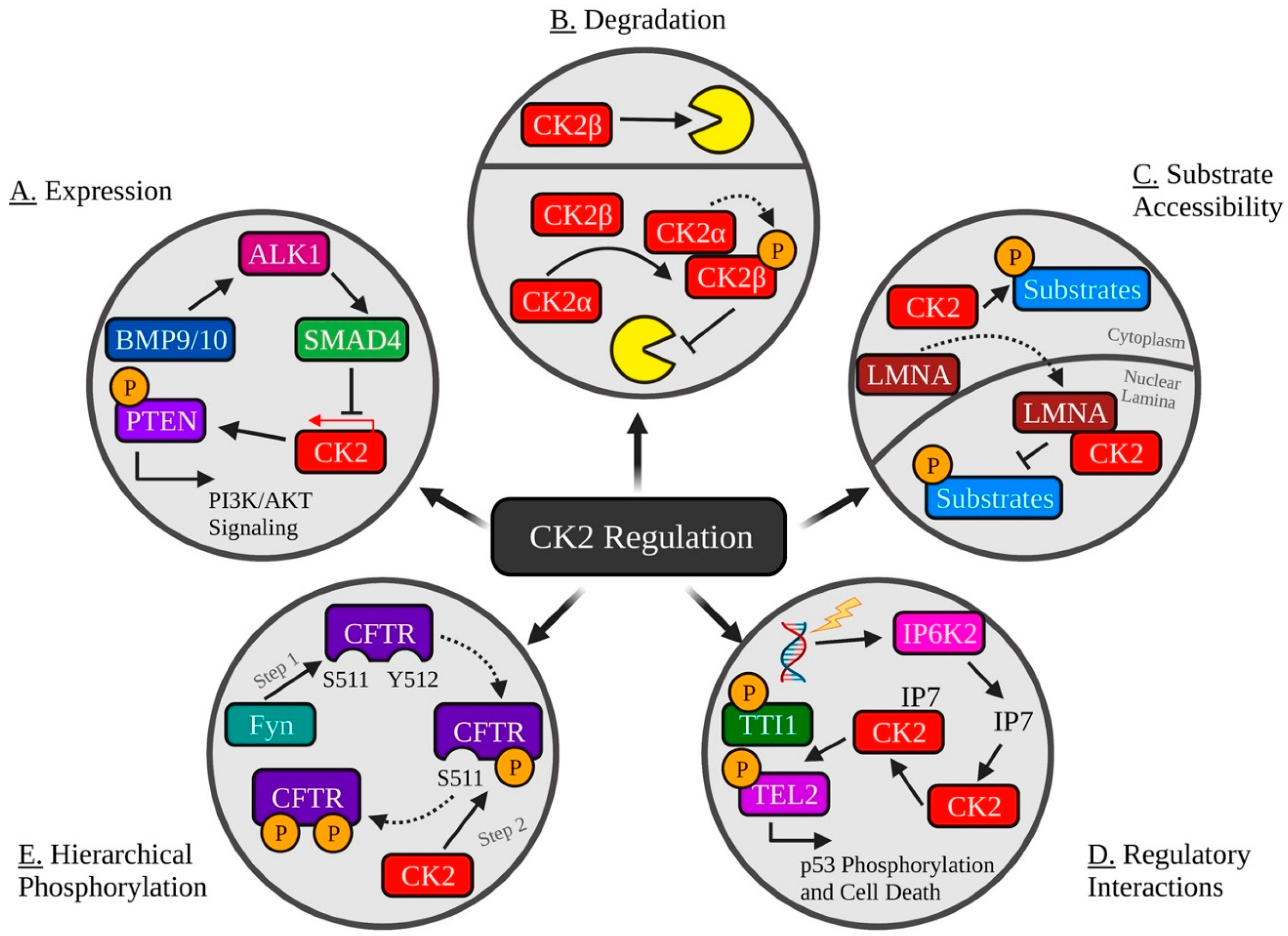
3. Extrinsic Regulation
3.1. Post-Translational Modifications
3.1.1. Activating
3.1.2. Inhibitory
3.2. Protein–Protein Interactions
3.2.1. Activating
3.2.2. Inhibitory
3.3. Regulatory Interactions with other Biological Molecules
3.3.1. Activating
3.3.2. Inhibitory
3.4. Hierarchical Phosphorylation
4. Unknown Regulatory Mechanism
4.1. Activating
4.2. Inhibitory
5. Summary and Perspectives
6. Concluding Remarks
Author Contributions
Funding
Conflicts of Interest
References
- Endicott, J.A.; Noble, M.E.; Johnson, L.N. The structural basis for control of eukaryotic protein kinases. Annu. Rev. Biochem. 2012, 81, 587–613. [Google Scholar] [CrossRef] [PubMed]
- Burnett, G.; Kennedy, E.P. The enzymatic phosphorylation of proteins. J. Biol. Chem. 1954, 211, 969–980. [Google Scholar] [CrossRef]
- Litchfield, D.W. Protein kinase CK2: Structure, regulation and role in cellular decisions of life and death. Biochem. J. 2003, 369, 1–15. [Google Scholar] [CrossRef]
- Beck, M.; Schmidt, A.; Malmstroem, J.; Claassen, M.; Ori, A.; Szymborska, A.; Herzog, F.; Rinner, O.; Ellenberg, J.; Aebersold, R. The quantitative proteome of a human cell line. Mol. Syst. Biol. 2011, 7, 549. [Google Scholar] [CrossRef] [PubMed]
- Geiger, T.; Wehner, A.; Schaab, C.; Cox, J.; Mann, M. Comparative proteomic analysis of eleven common cell lines reveals ubiquitous but varying expression of most proteins. Mol. Cell Proteom. 2012, 11, 9805–9808. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lundberg, E.; Fagerberg, L.; Klevebring, D.; Matic, I.; Geiger, T.; Cox, J.; Algenas, C.; Lundeberg, J.; Mann, M.; Uhlen, M. Defining the transcriptome and proteome in three functionally different human cell lines. Mol. Syst. Biol. 2010, 6, 450. [Google Scholar] [CrossRef]
- Poletto, G.; Vilardell, J.; Marin, O.; Pagano, M.A.; Cozza, G.; Sarno, S.; Falques, A.; Itarte, E.; Pinna, L.A.; Meggio, F. The regulatory beta subunit of protein kinase CK2 contributes to the recognition of the substrate consensus sequence. A study with an eIF2 beta-derived peptide. Biochemistry 2008, 47, 8317–8325. [Google Scholar] [CrossRef]
- UniProt, C. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef] [Green Version]
- Qaiser, F.; Trembley, J.H.; Kren, B.T.; Wu, J.J.; Naveed, A.K.; Ahmed, K. Protein kinase CK2 inhibition induces cell death via early impact on mitochondrial function. J. Cell Biochem. 2014, 115, 2103–2115. [Google Scholar] [CrossRef] [Green Version]
- Faust, M.; Jung, M.; Gunther, J.; Zimmermann, R.; Montenarh, M. Localization of individual subunits of protein kinase CK2 to the endoplasmic reticulum and to the Golgi apparatus. Mol. Cell Biochem. 2001, 227, 73–80. [Google Scholar] [CrossRef]
- Rodriguez, F.A.; Contreras, C.; Bolanos-Garcia, V.; Allende, J.E. Protein kinase CK2 as an ectokinase: The role of the regulatory CK2beta subunit. Proc. Natl. Acad. Sci. USA 2008, 105, 5693–5698. [Google Scholar] [CrossRef] [Green Version]
- Ponten, F.; Gry, M.; Fagerberg, L.; Lundberg, E.; Asplund, A.; Berglund, L.; Oksvold, P.; Bjorling, E.; Hober, S.; Kampf, C.; et al. A global view of protein expression in human cells, tissues, and organs. Mol. Syst. Biol. 2009, 5, 337. [Google Scholar] [CrossRef] [PubMed]
- Lou, D.Y.; Dominguez, I.; Toselli, P.; Landesman-Bollag, E.; O’Brien, C.; Seldin, D.C. The alpha catalytic subunit of protein kinase CK2 is required for mouse embryonic development. Mol. Cell Biol. 2008, 28, 131–139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buchou, T.; Vernet, M.; Blond, O.; Jensen, H.H.; Pointu, H.; Olsen, B.B.; Cochet, C.; Issinger, O.G.; Boldyreff, B. Disruption of the regulatory beta subunit of protein kinase CK2 in mice leads to a cell-autonomous defect and early embryonic lethality. Mol. Cell Biol. 2003, 23, 908–915. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, X.; Toselli, P.A.; Russell, L.D.; Seldin, D.C. Globozoospermia in mice lacking the casein kinase II alpha’ catalytic subunit. Nat. Genet 1999, 23, 118–121. [Google Scholar] [CrossRef] [PubMed]
- Meggio, F.; Pinna, L.A. One-thousand-and-one substrates of protein kinase CK2? FASEB J. 2003, 17, 349–368. [Google Scholar] [CrossRef] [PubMed]
- Zanin, S.; Sandre, M.; Cozza, G.; Ottaviani, D.; Marin, O.; Pinna, L.A.; Ruzzene, M. Chimeric peptides as modulators of CK2-dependent signaling: Mechanism of action and off-target effects. Biochim. Biophys. Acta 2015, 1854, 1694–1707. [Google Scholar] [CrossRef]
- Meggio, F.; Boldyreff, B.; Issinger, O.G.; Pinna, L.A. Casein kinase 2 down-regulation and activation by polybasic peptides are mediated by acidic residues in the 55-64 region of the beta-subunit. A study with calmodulin as phosphorylatable substrate. Biochemistry 1994, 33, 4336–4342. [Google Scholar] [CrossRef]
- Rosenberger, A.F.; Morrema, T.H.; Gerritsen, W.H.; van Haastert, E.S.; Snkhchyan, H.; Hilhorst, R.; Rozemuller, A.J.; Scheltens, P.; van der Vies, S.M.; Hoozemans, J.J. Increased occurrence of protein kinase CK2 in astrocytes in Alzheimer’s disease pathology. J. Neuroinflamm. 2016, 13, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castello, J.; Ragnauth, A.; Friedman, E.; Rebholz, H. CK2-An Emerging Target for Neurological and Psychiatric Disorders. Pharmaceuticals 2017, 10, 7. [Google Scholar] [CrossRef] [Green Version]
- Ampofo, E.; Nalbach, L.; Menger, M.D.; Montenarh, M.; Gotz, C. Protein Kinase CK2-A Putative Target for the Therapy of Diabetes Mellitus? Int. J. Mol. Sci. 2019, 20, 4398. [Google Scholar] [CrossRef] [Green Version]
- Baltan, S.; Bastian, C.; Quinn, J.; Aquila, D.; McCray, A.; Brunet, S. CK2 inhibition protects white matter from ischemic injury. Neurosci. Lett. 2018, 687, 37–42. [Google Scholar] [CrossRef]
- Ortega, C.E.; Seidner, Y.; Dominguez, I. Mining CK2 in cancer. PLoS ONE 2014, 9, e115609. [Google Scholar] [CrossRef]
- Borgo, C.; Ruzzene, M. Role of protein kinase CK2 in antitumor drug resistance. J. Exp. Clin. Cancer Res. 2019, 38, 287. [Google Scholar] [CrossRef]
- Siddiqui-Jain, A.; Drygin, D.; Streiner, N.; Chua, P.; Pierre, F.; O’Brien, S.E.; Bliesath, J.; Omori, M.; Huser, N.; Ho, C.; et al. CX-4945, an orally bioavailable selective inhibitor of protein kinase CK2, inhibits prosurvival and angiogenic signaling and exhibits antitumor efficacy. Cancer Res. 2010, 70, 10288–10298. [Google Scholar] [CrossRef] [Green Version]
- Perera, Y.; Ramos, Y.; Padron, G.; Caballero, E.; Guirola, O.; Caligiuri, L.G.; Lorenzo, N.; Gottardo, F.; Farina, H.G.; Filhol, O.; et al. CIGB-300 anticancer peptide regulates the protein kinase CK2-dependent phosphoproteome. Mol. Cell Biochem. 2020, 470, 63–75. [Google Scholar] [CrossRef]
- Yim, H.; Lee, Y.H.; Lee, C.H.; Lee, S.K. Emodin, an anthraquinone derivative isolated from the rhizomes of Rheum palmatum, selectively inhibits the activity of casein kinase II as a competitive inhibitor. Planta Med. 1999, 65, 9–13. [Google Scholar] [CrossRef]
- Lindenblatt, D.; Applegate, V.; Nickelsen, A.; Klussmann, M.; Neundorf, I.; Gotz, C.; Jose, J.; Niefind, K. Molecular Plasticity of Crystalline CK2alpha’ Leads to KN2, a Bivalent Inhibitor of Protein Kinase CK2 with Extraordinary Selectivity. J. Med. Chem. 2021. [Google Scholar] [CrossRef]
- Cozza, G.; Mazzorana, M.; Papinutto, E.; Bain, J.; Elliott, M.; di Maira, G.; Gianoncelli, A.; Pagano, M.A.; Sarno, S.; Ruzzene, M.; et al. Quinalizarin as a potent, selective and cell-permeable inhibitor of protein kinase CK2. Biochem. J. 2009, 421, 387–395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wells, C.I.; Drewry, D.H.; Pickett, J.E.; Tjaden, A.; Kramer, A.; Muller, S.; Gyenis, L.; Menyhart, D.; Litchfield, D.W.; Knapp, S.; et al. Development of a potent and selective chemical probe for the pleiotropic kinase CK2. Cell Chem. Biol. 2021, 28, 546–558.e10. [Google Scholar] [CrossRef] [PubMed]
- Duncan, J.S.; Gyenis, L.; Lenehan, J.; Bretner, M.; Graves, L.M.; Haystead, T.A.; Litchfield, D.W. An unbiased evaluation of CK2 inhibitors by chemoproteomics: Characterization of inhibitor effects on CK2 and identification of novel inhibitor targets. Mol. Cell Proteom. 2008, 7, 1077–1088. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okur, V.; Cho, M.T.; Henderson, L.; Retterer, K.; Schneider, M.; Sattler, S.; Niyazov, D.; Azage, M.; Smith, S.; Picker, J.; et al. De novo mutations in CSNK2A1 are associated with neurodevelopmental abnormalities and dysmorphic features. Hum. Genet 2016, 135, 699–705. [Google Scholar] [CrossRef] [PubMed]
- Akahira-Azuma, M.; Tsurusaki, Y.; Enomoto, Y.; Mitsui, J.; Kurosawa, K. Refining the clinical phenotype of Okur-Chung neurodevelopmental syndrome. Hum. Genome Var. 2018, 5, 18011. [Google Scholar] [CrossRef]
- Bouhaddou, M.; Memon, D.; Meyer, B.; White, K.M.; Rezelj, V.V.; Correa Marrero, M.; Polacco, B.J.; Melnyk, J.E.; Ulferts, S.; Kaake, R.M.; et al. The Global Phosphorylation Landscape of SARS-CoV-2 Infection. Cell 2020, 182, 685–712.e19. [Google Scholar] [CrossRef]
- Sarno, S.; Ghisellini, P.; Pinna, L.A. Unique activation mechanism of protein kinase CK2. The N-terminal segment is essential for constitutive activity of the catalytic subunit but not of the holoenzyme. J. Biol. Chem. 2002, 277, 22509–22514. [Google Scholar] [CrossRef] [Green Version]
- Donella-Deana, A.; Cesaro, L.; Sarno, S.; Brunati, A.M.; Ruzzene, M.; Pinna, L.A. Autocatalytic tyrosine-phosphorylation of protein kinase CK2 alpha and alpha’ subunits: Implication of Tyr182. Biochem. J. 2001, 357, 563–567. [Google Scholar] [CrossRef]
- Zhang, C.; Vilk, G.; Canton, D.A.; Litchfield, D.W. Phosphorylation regulates the stability of the regulatory CK2beta subunit. Oncogene 2002, 21, 3754–3764. [Google Scholar] [CrossRef] [Green Version]
- Krehan, A.; Ansuini, H.; Bocher, O.; Grein, S.; Wirkner, U.; Pyerin, W. Transcription factors ets1, NF-kappa B, and Sp1 are major determinants of the promoter activity of the human protein kinase CK2alpha gene. J. Biol. Chem. 2000, 275, 18327–18336. [Google Scholar] [CrossRef] [Green Version]
- Robitzki, A.; Bodenbach, L.; Voss, H.; Pyerin, W. Human casein kinase II. The subunit alpha protein activates transcription of the subunit beta gene. J. Biol. Chem. 1993, 268, 5694–5702. [Google Scholar] [CrossRef]
- Lolli, G.; Pinna, L.A.; Battistutta, R. Structural determinants of protein kinase CK2 regulation by autoinhibitory polymerization. ACS Chem. Biol. 2012, 7, 1158–1163. [Google Scholar] [CrossRef]
- Seetoh, W.G.; Chan, D.S.; Matak-Vinkovic, D.; Abell, C. Mass Spectrometry Reveals Protein Kinase CK2 High-Order Oligomerization via the Circular and Linear Assembly. ACS Chem. Biol. 2016, 11, 1511–1517. [Google Scholar] [CrossRef] [Green Version]
- Hubner, G.M.; Larsen, J.N.; Guerra, B.; Niefind, K.; Vrecl, M.; Issinger, O.G. Evidence for aggregation of protein kinase CK2 in the cell: A novel strategy for studying CK2 holoenzyme interaction by BRET(2). Mol. Cell Biochem. 2014, 397, 285–293. [Google Scholar] [CrossRef]
- Lolli, G.; Ranchio, A.; Battistutta, R. Active form of the protein kinase CK2 alpha2beta2 holoenzyme is a strong complex with symmetric architecture. ACS Chem. Biol. 2014, 9, 366–371. [Google Scholar] [CrossRef] [PubMed]
- Schnitzler, A.; Olsen, B.B.; Issinger, O.G.; Niefind, K. The protein kinase CK2(Andante) holoenzyme structure supports proposed models of autoregulation and trans-autophosphorylation. J. Mol. Biol. 2014, 426, 1871–1882. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, B.S. Akt activation enhances ribosomal RNA synthesis through casein kinase II and TIF-IA. Proc. Natl. Acad. Sci. USA 2013, 110, 20681–20686. [Google Scholar] [CrossRef] [Green Version]
- Ji, H.; Wang, J.; Nika, H.; Hawke, D.; Keezer, S.; Ge, Q.; Fang, B.; Fang, X.; Fang, D.; Litchfield, D.W.; et al. EGF-induced ERK activation promotes CK2-mediated disassociation of alpha-Catenin from beta-Catenin and transactivation of beta-Catenin. Mol. Cell 2009, 36, 547–559. [Google Scholar] [CrossRef] [Green Version]
- Donella-Deana, A.; Cesaro, L.; Sarno, S.; Ruzzene, M.; Brunati, A.M.; Marin, O.; Vilk, G.; Doherty-Kirby, A.; Lajoie, G.; Litchfield, D.W.; et al. Tyrosine phosphorylation of protein kinase CK2 by Src-related tyrosine kinases correlates with increased catalytic activity. Biochem. J. 2003, 372, 841–849. [Google Scholar] [CrossRef] [Green Version]
- Goel, R.K.; Meyer, M.; Paczkowska, M.; Reimand, J.; Vizeacoumar, F.; Vizeacoumar, F.; Lam, T.T.; Lukong, K.E. Global phosphoproteomic analysis identifies SRMS-regulated secondary signaling intermediates. Proteome Sci. 2018, 16, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, Y.H.; Park, J.W.; Bae, Y.S. Regulation of protein kinase CK2 catalytic activity by protein kinase C and phospholipase D2. Biochimie 2016, 121, 131–139. [Google Scholar] [CrossRef] [PubMed]
- St-Denis, N.A.; Derksen, D.R.; Litchfield, D.W. Evidence for regulation of mitotic progression through temporal phosphorylation and dephosphorylation of CK2alpha. Mol. Cell Biol. 2009, 29, 2068–2081. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Litchfield, D.W.; Luscher, B.; Lozeman, F.J.; Eisenman, R.N.; Krebs, E.G. Phosphorylation of casein kinase II by p34cdc2 in vitro and at mitosis. J. Biol. Chem. 1992, 267, 13943–13951. [Google Scholar] [CrossRef]
- Litchfield, D.W.; Lozeman, F.J.; Cicirelli, M.F.; Harrylock, M.; Ericsson, L.H.; Piening, C.J.; Krebs, E.G. Phosphorylation of the beta subunit of casein kinase II in human A431 cells. Identification of the autophosphorylation site and a site phosphorylated by p34cdc2. J. Biol. Chem. 1991, 266, 20380–20389. [Google Scholar] [CrossRef]
- Bosc, D.G.; Luscher, B.; Litchfield, D.W. Expression and regulation of protein kinase CK2 during the cell cycle. Mol. Cell Biochem. 1999, 191, 213–222. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Ham, S.; Yang, K.; Kim, K. Protein kinase CK2 activation is required for transforming growth factor beta-induced epithelial-mesenchymal transition. Mol. Oncol. 2018, 12, 1811–1826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choudhary, C.; Kumar, C.; Gnad, F.; Nielsen, M.L.; Rehman, M.; Walther, T.C.; Olsen, J.V.; Mann, M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 2009, 325, 834–840. [Google Scholar] [CrossRef] [Green Version]
- Huang, R.; Holbert, M.A.; Tarrant, M.K.; Curtet, S.; Colquhoun, D.R.; Dancy, B.M.; Dancy, B.C.; Hwang, Y.; Tang, Y.; Meeth, K.; et al. Site-specific introduction of an acetyl-lysine mimic into peptides and proteins by cysteine alkylation. J. Am. Chem. Soc. 2010, 132, 9986–9987. [Google Scholar] [CrossRef] [Green Version]
- Tarrant, M.K.; Rho, H.S.; Xie, Z.; Jiang, Y.L.; Gross, C.; Culhane, J.C.; Yan, G.; Qian, J.; Ichikawa, Y.; Matsuoka, T.; et al. Regulation of CK2 by phosphorylation and O-GlcNAcylation revealed by semisynthesis. Nat. Chem. Biol. 2012, 8, 262–269. [Google Scholar] [CrossRef] [Green Version]
- Yao, Q.; Li, H.; Liu, B.Q.; Huang, X.Y.; Guo, L. SUMOylation-regulated protein phosphorylation, evidence from quantitative phosphoproteomics analyses. J. Biol. Chem. 2011, 286, 27342–27349. [Google Scholar] [CrossRef] [Green Version]
- Skjerpen, C.S.; Nilsen, T.; Wesche, J.; Olsnes, S. Binding of FGF-1 variants to protein kinase CK2 correlates with mitogenicity. EMBO J. 2002, 21, 4058–4069. [Google Scholar] [CrossRef] [Green Version]
- Bonnet, H.; Filhol, O.; Truchet, I.; Brethenou, P.; Cochet, C.; Amalric, F.; Bouche, G. Fibroblast growth factor-2 binds to the regulatory beta subunit of CK2 and directly stimulates CK2 activity toward nucleolin. J. Biol. Chem. 1996, 271, 24781–24787. [Google Scholar] [CrossRef] [Green Version]
- Yu, S.; Li, L.; Cai, H.; He, B.; Gao, Y.; Li, Y. Overexpression of NELFE contributes to gastric cancer progression via Wnt/beta-catenin signaling-mediated activation of CSNK2B expression. J. Exp. Clin. Cancer Res. 2021, 40, 54. [Google Scholar] [CrossRef] [PubMed]
- Plotnikov, A.; Chuderland, D.; Karamansha, Y.; Livnah, O.; Seger, R. Nuclear ERK Translocation is Mediated by Protein Kinase CK2 and Accelerated by Autophosphorylation. Cell Physiol. Biochem. 2019, 53, 366–387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, Z.M.; Ai, X.; Teng, J.F.; Wang, Y.P.; Wang, B.J.; Zhang, X. p21 and CK2 interaction-mediated HDAC2 phosphorylation modulates KLF4 acetylation to regulate bladder cancer cell proliferation. Tumour. Biol. 2016, 37, 8293–8304. [Google Scholar] [CrossRef] [PubMed]
- Gotz, C.; Wagner, P.; Issinger, O.G.; Montenarh, M. p21WAF1/CIP1 interacts with protein kinase CK2. Oncogene 1996, 13, 391–398. [Google Scholar]
- Yang, Y.; Fan, X.; Ren, Y.; Wu, K.; Tian, X.; Wen, F.; Liu, D.; Fan, Y.; Zhao, S. SOX2-Upregulated microRNA-30e Promotes the Progression of Esophageal Cancer via Regulation of the USP4/SMAD4/CK2 Axis. Mol. Ther. Nucleic Acids 2021, 23, 200–214. [Google Scholar] [CrossRef]
- Arriazu, E.; Vicente, C.; Pippa, R.; Peris, I.; Martinez-Balsalobre, E.; Garcia-Ramirez, P.; Marcotegui, N.; Igea, A.; Alignani, D.; Rifon, J.; et al. A new regulatory mechanism of protein phosphatase 2A activity via SET in acute myeloid leukemia. Blood Cancer J. 2020, 10, 3. [Google Scholar] [CrossRef]
- St-Denis, N.A.; Bailey, M.L.; Parker, E.L.; Vilk, G.; Litchfield, D.W. Localization of phosphorylated CK2alpha to the mitotic spindle requires the peptidyl-prolyl isomerase Pin1. J. Cell Sci. 2011, 124, 2341–2348. [Google Scholar] [CrossRef] [Green Version]
- Chen, T.; Chen, J.; Zhu, Y.; Li, Y.; Wang, Y.; Chen, H.; Wang, J.; Li, X.; Liu, Y.; Li, B.; et al. CD163, a novel therapeutic target, regulates the proliferation and stemness of glioma cells via casein kinase 2. Oncogene 2019, 38, 1183–1199. [Google Scholar] [CrossRef]
- Raman, C.; Kuo, A.; Deshane, J.; Litchfield, D.W.; Kimberly, R.P. Regulation of casein kinase 2 by direct interaction with cell surface receptor CD5. J. Biol. Chem. 1998, 273, 19183–19189. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.B.; Shin, Y.J.; Roy, A.; Kim, J.H. The Role of the Pleckstrin Homology Domain-containing Protein CKIP-1 in Activation of p21-activated Kinase 1 (PAK1). J. Biol. Chem. 2015, 290, 21076–21085. [Google Scholar] [CrossRef] [Green Version]
- Bosc, D.G.; Graham, K.C.; Saulnier, R.B.; Zhang, C.; Prober, D.; Gietz, R.D.; Litchfield, D.W. Identification and characterization of CKIP-1, a novel pleckstrin homology domain-containing protein that interacts with protein kinase CK2. J. Biol. Chem. 2000, 275, 14295–14306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Litchfield, D.W.; Bosc, D.G.; Canton, D.A.; Saulnier, R.B.; Vilk, G.; Zhang, C. Functional specialization of CK2 isoforms and characterization of isoform-specific binding partners. Mol. Cell Biochem. 2001, 227, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Olsten, M.E.; Canton, D.A.; Zhang, C.; Walton, P.A.; Litchfield, D.W. The Pleckstrin homology domain of CK2 interacting protein-1 is required for interactions and recruitment of protein kinase CK2 to the plasma membrane. J. Biol. Chem. 2004, 279, 42114–42127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keller, D.M.; Zeng, X.; Wang, Y.; Zhang, Q.H.; Kapoor, M.; Shu, H.; Goodman, R.; Lozano, G.; Zhao, Y.; Lu, H. A DNA damage-induced p53 serine 392 kinase complex contains CK2, hSpt16, and SSRP1. Mol. Cell 2001, 7, 283–292. [Google Scholar] [CrossRef]
- Miyata, Y.; Yahara, I. Interaction between casein kinase II and the 90-kDa stress protein, HSP90. Biochemistry 1995, 34, 8123–8129. [Google Scholar] [CrossRef]
- Chen, Q.; Hao, W.; Xiao, C.; Wang, R.; Xu, X.; Lu, H.; Chen, W.; Deng, C.X. SIRT6 Is Essential for Adipocyte Differentiation by Regulating Mitotic Clonal Expansion. Cell Rep. 2017, 18, 3155–3166. [Google Scholar] [CrossRef]
- Wang, Y.; Schachner, M. The intracellular domain of L1CAM binds to casein kinase 2alpha and is neuroprotective via inhibition of the tumor suppressors PTEN and p53. J. Neurochem. 2015, 133, 828–843. [Google Scholar] [CrossRef]
- Ao, Y.; Zhang, J.; Liu, Z.; Qian, M.; Li, Y.; Wu, Z.; Sun, P.; Wu, J.; Bei, W.; Wen, J.; et al. Lamin A buffers CK2 kinase activity to modulate aging in a progeria mouse model. Sci. Adv. 2019, 5, eaav5078. [Google Scholar] [CrossRef] [Green Version]
- Messenger, M.M.; Saulnier, R.B.; Gilchrist, A.D.; Diamond, P.; Gorbsky, G.J.; Litchfield, D.W. Interactions between protein kinase CK2 and Pin1. Evidence for phosphorylation-dependent interactions. J. Biol. Chem. 2002, 277, 23054–23064. [Google Scholar] [CrossRef] [Green Version]
- Ola, R.; Kunzel, S.H.; Zhang, F.; Genet, G.; Chakraborty, R.; Pibouin-Fragner, L.; Martin, K.; Sessa, W.; Dubrac, A.; Eichmann, A. SMAD4 Prevents Flow Induced Arteriovenous Malformations by Inhibiting Casein Kinase 2. Circulation 2018, 138, 2379–2394. [Google Scholar] [CrossRef]
- Xiao, Y.; Huang, S.; Qiu, F.; Ding, X.; Sun, Y.; Wei, C.; Hu, X.; Wei, K.; Long, S.; Xie, L.; et al. Tumor necrosis factor alpha-induced protein 1 as a novel tumor suppressor through selective downregulation of CSNK2B blocks nuclear factor-kappaB activation in hepatocellular carcinoma. EBioMedicine 2020, 51, 102603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanz-Clemente, A.; Gray, J.A.; Ogilvie, K.A.; Nicoll, R.A.; Roche, K.W. Activated CaMKII couples GluN2B and casein kinase 2 to control synaptic NMDA receptors. Cell Rep. 2013, 3, 607–614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patel, S.; Rauf, A.; Khan, H.; Abu-Izneid, T. Renin-angiotensin-aldosterone (RAAS): The ubiquitous system for homeostasis and pathologies. Biomed. Pharmacother. 2017, 94, 317–325. [Google Scholar] [CrossRef]
- Kashihara, T.; Nakada, T.; Kojima, K.; Takeshita, T.; Yamada, M. Angiotensin II activates CaV 1.2 Ca(2+) channels through beta-arrestin2 and casein kinase 2 in mouse immature cardiomyocytes. J. Physiol. 2017, 595, 4207–4225. [Google Scholar] [CrossRef] [Green Version]
- McGuire, D.J.; Rowse, A.L.; Li, H.; Peng, B.J.; Sestero, C.M.; Cashman, K.S.; De Sarno, P.; Raman, C. CD5 enhances Th17-cell differentiation by regulating IFN-gamma response and RORgammat localization. Eur. J. Immunol. 2014, 44, 1137–1142. [Google Scholar] [CrossRef] [Green Version]
- di Masi, A.; De Simone, G.; Ciaccio, C.; D’Orso, S.; Coletta, M.; Ascenzi, P. Haptoglobin: From hemoglobin scavenging to human health. Mol. Asp. Med. 2020, 73, 100851. [Google Scholar] [CrossRef]
- Homma, M.K.; Li, D.; Krebs, E.G.; Yuasa, Y.; Homma, Y. Association and regulation of casein kinase 2 activity by adenomatous polyposis coli protein. Proc. Natl. Acad. Sci. USA 2002, 99, 5959–5964. [Google Scholar] [CrossRef] [Green Version]
- Leroy, D.; Filhol, O.; Delcros, J.G.; Pares, S.; Chambaz, E.M.; Cochet, C. Chemical features of the protein kinase CK2 polyamine binding site. Biochemistry 1997, 36, 1242–1250. [Google Scholar] [CrossRef]
- Leroy, D.; Schmid, N.; Behr, J.P.; Filhol, O.; Pares, S.; Garin, J.; Bourgarit, J.J.; Chambaz, E.M.; Cochet, C. Direct identification of a polyamine binding domain on the regulatory subunit of the protein kinase casein kinase 2 by photoaffinity labeling. J. Biol. Chem. 1995, 270, 17400–17406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leroy, D.; Heriche, J.K.; Filhol, O.; Chambaz, E.M.; Cochet, C. Binding of polyamines to an autonomous domain of the regulatory subunit of protein kinase CK2 induces a conformational change in the holoenzyme. A proposed role for the kinase stimulation. J. Biol. Chem. 1997, 272, 20820–20827. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shore, L.J.; Soler, A.P.; Gilmour, S.K. Ornithine decarboxylase expression leads to translocation and activation of protein kinase CK2 in vivo. J. Biol. Chem. 1997, 272, 12536–12543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, R.; Al-Quobaili, F.; Muller, I.; Gotz, C.; Thiel, G.; Montenarh, M. CK2 phosphorylation of Pdx-1 regulates its transcription factor activity. Cell Mol. Life Sci. 2010, 67, 2481–2489. [Google Scholar] [CrossRef] [PubMed]
- Meng, R.; Gotz, C.; Montenarh, M. The role of protein kinase CK2 in the regulation of the insulin production of pancreatic islets. Biochem. Biophys. Res. Commun. 2010, 401, 203–206. [Google Scholar] [CrossRef] [PubMed]
- Klein, S.; Meng, R.; Montenarh, M.; Gotz, C. The Phosphorylation of PDX-1 by Protein Kinase CK2 Is Crucial for Its Stability. Pharmaceuticals 2016, 10, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abu Shehab, M.; Biggar, K.; Kakadia, J.H.; Dhruv, M.; Jain, B.; Nandi, P.; Nygard, K.; Jansson, T.; Gupta, M.B. Inhibition of decidual IGF-1 signaling in response to hypoxia and leucine deprivation is mediated by mTOR and AAR pathways and increased IGFBP-1 phosphorylation. Mol. Cell Endocrinol. 2020, 512, 110865. [Google Scholar] [CrossRef]
- Pitolli, C.; Wang, Y.; Candi, E.; Shi, Y.; Melino, G.; Amelio, I. p53-Mediated Tumor Suppression: DNA-Damage Response and Alternative Mechanisms. Cancers 2019, 11, 1983. [Google Scholar] [CrossRef] [Green Version]
- Rao, F.; Cha, J.; Xu, J.; Xu, R.; Vandiver, M.S.; Tyagi, R.; Tokhunts, R.; Koldobskiy, M.A.; Fu, C.; Barrow, R.; et al. Inositol pyrophosphates mediate the DNA-PK/ATM-p53 cell death pathway by regulating CK2 phosphorylation of Tti1/Tel2. Mol. Cell 2014, 54, 119–132. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Wang, Y.; Xu, M.; Zhang, L.; Su, Y.; Wang, B.; Zhang, X. miR-1184 regulates the proliferation and apoptosis of colon cancer cells via targeting CSNK2A1. Mol. Cell Probes 2020, 53, 101625. [Google Scholar] [CrossRef]
- Oduah, E.I.; Linhardt, R.J.; Sharfstein, S.T. Heparin: Past, Present, and Future. Pharmaceuticals 2016, 9, 38. [Google Scholar] [CrossRef]
- O’Farrell, F.; Loog, M.; Janson, I.M.; Ek, P. Kinetic study of the inhibition of CK2 by heparin fragments of different length. Biochim. Biophys. Acta 1999, 1433, 68–75. [Google Scholar] [CrossRef]
- Top, D.; Harms, E.; Syed, S.; Adams, E.L.; Saez, L. GSK-3 and CK2 Kinases Converge on Timeless to Regulate the Master Clock. Cell Rep. 2016, 16, 357–367. [Google Scholar] [CrossRef] [Green Version]
- Venerando, A.; Cesaro, L.; Marin, O.; Donella-Deana, A.; Pinna, L.A. A “SYDE” effect of hierarchical phosphorylation: Possible relevance to the cystic fibrosis basic defect. Cell Mol. Life Sci. 2014, 71, 2193–2196. [Google Scholar] [CrossRef] [PubMed]
- Cesaro, L.; Marin, O.; Venerando, A.; Donella-Deana, A.; Pinna, L.A. Phosphorylation of cystic fibrosis transmembrane conductance regulator (CFTR) serine-511 by the combined action of tyrosine kinases and CK2: The implication of tyrosine-512 and phenylalanine-508. Amino Acids 2013, 45, 1423–1429. [Google Scholar] [CrossRef] [PubMed]
- Marin, O.; Meggio, F.; Perich, J.W.; Pinna, L.A. Phosphotyrosine specifies the phosphorylation by protein kinase CK2 of a peptide reproducing the activation loop of the insulin receptor protein tyrosine kinase. Int. J. Biochem. Cell Biol. 1996, 28, 999–1005. [Google Scholar] [CrossRef]
- Hoque, A.; Williamson, N.A.; Ameen, S.S.; Ciccotosto, G.D.; Hossain, M.I.; Oakhill, J.S.; Ng, D.C.H.; Ang, C.S.; Cheng, H.C. Quantitative proteomic analyses of dynamic signalling events in cortical neurons undergoing excitotoxic cell death. Cell Death Dis. 2019, 10, 213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, A.K.; Ahn, S.G.; Yoon, J.H.; Kim, S.A. Sox4 stimulates ß-catenin activity through induction of CK2. Oncol. Rep. 2011, 25, 559–565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Posch, C.; Sanlorenzo, M.; Vujic, I.; Oses-Prieto, J.A.; Cholewa, B.D.; Kim, S.T.; Ma, J.; Lai, K.; Zekhtser, M.; Esteve-Puig, R.; et al. Phosphoproteomic Analyses of NRAS(G12) and NRAS(Q61) Mutant Melanocytes Reveal Increased CK2alpha Kinase Levels in NRAS(Q61) Mutant Cells. J. Investig. Dermatol. 2016, 136, 2041–2048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Basu, S.K.; Lee, S.; Salotti, J.; Basu, S.; Sakchaisri, K.; Xiao, Z.; Walia, V.; Westlake, C.J.; Morrison, D.K.; Johnson, P.F. Oncogenic RAS-Induced Perinuclear Signaling Complexes Requiring KSR1 Regulate Signal Transmission to Downstream Targets. Cancer Res. 2018, 78, 891–908. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dompe, C.; Moncrieff, L.; Matys, J.; Grzech-Lesniak, K.; Kocherova, I.; Bryja, A.; Bruska, M.; Dominiak, M.; Mozdziak, P.; Skiba, T.H.I.; et al. Photobiomodulation-Underlying Mechanism and Clinical Applications. J. Clin. Med. 2020, 9, 1724. [Google Scholar] [CrossRef]
- Chang, L.Y.; Fan, S.M.; Liao, Y.C.; Wang, W.H.; Chen, Y.J.; Lin, S.J. Proteomic Analysis Reveals Anti-Fibrotic Effects of Blue Light Photobiomodulation on Fibroblasts. Lasers Surg Med. 2020, 52, 358–372. [Google Scholar] [CrossRef]
- Hardesty, J.E.; Wahlang, B.; Falkner, K.C.; Shi, H.; Jin, J.; Wilkey, D.; Merchant, M.; Watson, C.; Prough, R.A.; Cave, M.C. Hepatic signalling disruption by pollutant Polychlorinated biphenyls in steatohepatitis. Cell Signal 2019, 53, 132–139. [Google Scholar] [CrossRef] [PubMed]
- Nagahori, H.; Nakamura, K.; Sumida, K.; Ito, S.; Ohtsuki, S. Combining Genomics To Identify the Pathways of Post-Transcriptional Nongenotoxic Signaling and Energy Homeostasis in Livers of Rats Treated with the Pregnane X Receptor Agonist, Pregnenolone Carbonitrile. J. Proteome Res. 2017, 16, 3634–3645. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Yu, S.; Davis, A.T.; Ahmed, K. Cell cycle dependent regulation of protein kinase CK2 signaling to the nuclear matrix. J. Cell Biochem. 2003, 88, 812–822. [Google Scholar] [CrossRef]
- Homma, M.K.; Homma, Y. Cell cycle and activation of CK2. Mol. Cell Biochem. 2008, 316, 49–55. [Google Scholar] [CrossRef] [PubMed]
| Inhibitor | Classification | Mechanism | In Clinical Trials? | Refs. |
|---|---|---|---|---|
| CIGB-300 | Peptide | Direct Substrate Binding | Yes | [26] |
| Emodin | Natural Product | ATP Competitive | Yes 1 | [27] |
| KN2 | Small Molecule | Bivalent Inhibition | No | [28] |
| Quinalizarin | Small Molecule | ATP Competitive | No | [29] |
| SGC-CK2-1 | Small Molecule | ATP Competitive | No | [30] |
| Silmitasertib | Small Molecule | ATP Competitive | Yes | [25] |
| TBB | Small Molecule | ATP Competitive | No | [31] |
| TBBz | Small Molecule | ATP Competitive | No | [31] |
| Interactor | Effect on CK2 Activity | Affected CK2 Substrate(s) | Biological Function | Refs. |
|---|---|---|---|---|
| APC | Inhibitory | CK2⍺/CK2β | Cell Proliferation | [65] |
| AT2 | Activating | Cav1.2 | Cardiovascular Regulation | [66] |
| CaMKII | Activating | GluN2B | Glutamate Signalling | [67] |
| CD163 | Activating | AKT | Cell Proliferation | [68] |
| CD5 | Activating | N/A | Adaptive Immunity | [69] |
| CKIP-1 | Activating | PAK1 | Cell Morphology | [70,71,72,73] |
| FACT | Activating | p53 | DNA Damage Response | [74] |
| FGF-1 | Activating | CK2β | Cell Proliferation | [59] |
| FGF-2 | Activating | Nucleolin | Cell Proliferation | [60] |
| HSP90 | Activating | N/A | Stress Response | [75] |
| KIF5C | Inhibitory | N/A | Cell Cycle | [76] |
| L1CAM | Activating | PTEN; p53 | Neuritogenesis | [77] |
| Lamin A | Inhibitory | N/A | Cellular Senescence | [78] |
| MEK | Activating | ERK | Cell Proliferation | [62] |
| NELFE | Activating | N/A | Cell Proliferation | [61] |
| p21 | Activating | HDAC2 | Cell Proliferation | [63,64] |
| p38β | Activating | SET | Stress Response | [66] |
| Pin1 | Activating | N/A | Cell Cycle | [67] |
| Pin1 | Inhibitory | TOP2A | Cell Cycle | [79] |
| SMAD4 | Inhibitory | PTEN | Cell Proliferation | [80] |
| SOX2 | Activating | N/A | Cell Proliferation | [65] |
| TNFAIP1 | Inhibitory | N/A | Cell Proliferation | [81] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roffey, S.E.; Litchfield, D.W. CK2 Regulation: Perspectives in 2021. Biomedicines 2021, 9, 1361. https://0-doi-org.brum.beds.ac.uk/10.3390/biomedicines9101361
Roffey SE, Litchfield DW. CK2 Regulation: Perspectives in 2021. Biomedicines. 2021; 9(10):1361. https://0-doi-org.brum.beds.ac.uk/10.3390/biomedicines9101361
Chicago/Turabian StyleRoffey, Scott E., and David W. Litchfield. 2021. "CK2 Regulation: Perspectives in 2021" Biomedicines 9, no. 10: 1361. https://0-doi-org.brum.beds.ac.uk/10.3390/biomedicines9101361






