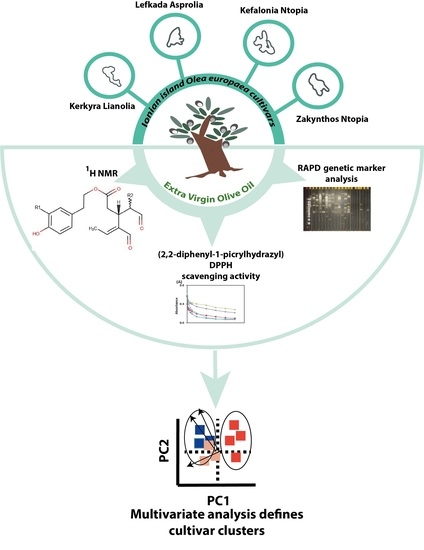
A Study on the Clustering of Extra Virgin Olive Oils Extracted from Cultivars Growing in Four Ionian Islands (Greece) by Multivariate Analysis of Their Phenolic Profile, Antioxidant Activity and Genetic Markers
Abstract
:1. Introduction
2. Materials and Methods
2.1. Olive Fruit Collection and Olive Oil Extraction Process
2.2. Sample Preparation for NMR Analysis
2.3. NMR Spectral Analysis
2.4. Determination of Antioxidant Activity
2.5. DNA Extraction
2.6. Random Amplified Polymorphic DNA (RAPDS)
2.7. Genetic Distance Estimation
2.8. Multivariate Statistical Analyses
3. Results and Discussion
3.1. BioPhenolic Compound Concentrations of Olive Oils from Ionian Island Cultivars
3.2. Cultivar Effect on the Relationship between Olive Oil Antioxidant Activity and Individual Secoiridoids
3.3. Multivariate Analysis of the Phenolic Profile Clusters Samples by Cultivar Geographic Origin
3.4. Ionian Island Cultivar Genetic Diversity Based on RAPD Genetic Markers
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAOSTAT Data 2018. Available online: https://www.fao.org/faostat/en/#data/QCL (accessed on 21 November 2021).
- Romani, A.; Ieri, F.; Urciuoli, S.; Noce, A.; Marrone, G.; Nediani, C.; Bernini, R. Health Effects of Phenolic Compounds Found in Extra-Virgin Olive Oil, By-Products, and Leaf of Olea europaea L. Nutrients 2019, 11, 1776. [Google Scholar] [CrossRef] [Green Version]
- Tripoli, E.; Giammanco, M.; Tabacchi, G.; Di Majo, D.; Giammanco, S.; La Guardia, M. The phenolic compounds of olive oil: Structure, biological activity and beneficial effects on human health. Nutr. Res. Rev. 2005, 18, 98–112. [Google Scholar] [CrossRef] [PubMed]
- Gorzynik-Debicka, M.; Przychodzen, P.; Cappello, F.; Kuban-Jankowska, A.; Marino Gammazza, A.; Knap, N.; Wozniak, M.; Gorska-Ponikowska, M. Potential Health Benefits of Olive Oil and Plant Polyphenols. Int. J. Mol. Sci. 2018, 19, 686. [Google Scholar] [CrossRef] [Green Version]
- Rahmani, A.H.; Albutti, A.S.; Aly, S.M. Therapeutics role of olive fruits/oil in the prevention of diseases via modulation of anti-oxidant, anti-tumour and genetic activity. Int. J. Clin. Exp. Med. 2014, 7, 799–808. [Google Scholar] [PubMed]
- Rigacci, S.; Stefani, M. Nutraceutical Properties of Olive Oil Polyphenols. An Itinerary from Cultured Cells through Animal Models to Humans. Int. J. Mol. Sci. 2016, 17, 843. [Google Scholar] [CrossRef] [Green Version]
- Tsolaki, M.; Lazarou, E.; Kozori, M.; Petridou, N.; Tabakis, I.; Lazarou, I.; Karakota, M.; Saoulidis, I.; Melliou, E.; Magiatis, P. A Randomized Clinical Trial of Greek High Phenolic Early Harvest Extra Virgin Olive Oil in Mild Cognitive Impairment: The MICOIL Pilot Study. J. Alzheimer’s Dis. 2020, 78, 801–817. [Google Scholar] [CrossRef]
- Schwingshackl, L.; Hoffmann, G. Monounsaturated fatty acids, olive oil and health status: A systematic review and meta-analysis of cohort studies. Lipids Health Dis. 2014, 13. [Google Scholar] [CrossRef] [Green Version]
- Jimenez-Lopez, C.; Carpena, M.; Lourenço-Lopes, C.; Gallardo-Gomez, M.; Lorenzo, J.M.; Barba, F.J.; Prieto, M.A.; Simal-Gandara, J. Bioactive Compounds and Quality of Extra Virgin Olive Oil. Foods 2020, 9, 1014. [Google Scholar] [CrossRef] [PubMed]
- Stark, A.H.; Madar, Z. Olive oil as a functional food: Epidemiology and nutritional approaches. Nutr. Rev. 2002, 60, 170–176. [Google Scholar] [CrossRef]
- Aparicio-Soto, M.; Sánchez-Hidalgo, M.; Rosillo, M.Á.; Castejón, M.L.; Alarcón-De-La-Lastra, C. Extra virgin olive oil: A key functional food for prevention of immune-inflammatory diseases. Food Funct. 2016, 7, 4492–4505. [Google Scholar] [CrossRef]
- EFSA. Scientific Opinion on the substantiation of health claims related to the replacement of mixtures of saturated fatty acids (SFAs) as present in foods or diets with mixtures of monounsaturated fatty acids (MUFAs) and/or mixtures of polyunsaturated fatty acids (PUFAs), and maintenance of normal blood LDL-cholesterol concentrations. EFSA J. 2011, 9, 2069. [Google Scholar]
- De La Torre-Carbot, K.; Chávez-Servín, J.L.; Jaúregui, O.; Castellote-Bargalló, A.I.; Lamuela-Raventos, R.M.; Nurmi, T.; Poulsen, H.E.; Gaddi, A.V.; Kaikkonen, J.; Zunft, H.-F.; et al. Elevated Circulating LDL Phenol Levels in Men Who Consumed Virgin Rather Than Refined Olive Oil Are Associated with Less Oxidation of Plasma LDL. J. Nutr. 2010, 140, 501–508. [Google Scholar] [CrossRef]
- Karkoula, E.; Skantzari, A.; Melliou, E.; Magiatis, P. Direct Measurement of Oleocanthal and Oleacein Levels in Olive Oil by Quantitative 1H NMR. Establishment of a New Index for the Characterization of Extra Virgin Olive Oils. J. Agric. Food Chem. 2012, 60, 11696–11703. [Google Scholar] [CrossRef]
- Karkoula, E.; Skantzari, A.; Melliou, E.; Magiatis, P. Quantitative Measurement of Major Secoiridoid Derivatives in Olive Oil Using qNMR. Proof of the Artificial Formation of Aldehydic Oleuropein and Ligstroside Aglycon Isomers. J. Agric. Food Chem. 2014, 62, 600–607. [Google Scholar] [CrossRef]
- Diamantakos, P.; Velkou, A.; Killday, B.K.; Gimisis, T.; Melliou, E.; Magiatis, P. Oleokoronal and oleomissional: New major phenolic ingredients of extra virgin olive oil. Olivae 2015, 122, 22–33. [Google Scholar]
- Angelis, A.; Antoniadi, L.; Stathopoulos, P.; Halabalaki, M.; Skaltsounis, L.A. Oleocanthalic and Oleaceinic acids: New compounds from Extra Virgin Olive Oil (EVOO). Phytochem. Lett. 2018, 26, 190–194. [Google Scholar] [CrossRef]
- Tsolakou, A.; Diamantakos, P.; Kalaboki, I.; Mena-Bravo, A.; Priego-Capote, F.; Abdallah, I.M.; Kaddoumi, A.; Melliou, E.; Magiatis, P. Oleocanthalic Acid, a Chemical Marker of Olive Oil Aging and Exposure to a High Storage Temperature with Potential Neuroprotective Activity. J. Agric. Food Chem. 2018, 66, 7337–7346. [Google Scholar] [CrossRef]
- Bellumori, M.; Cecchi, L.; Innocenti, M.; Clodoveo, M.L.; Corbo, F.; Mulinacci, N. The EFSA Health Claim on Olive Oil Polyphenols: Acid Hydrolysis Validation and Total Hydroxytyrosol and Tyrosol Determination in Italian Virgin Olive Oils. Molecules 2019, 24, 2179. [Google Scholar] [CrossRef] [Green Version]
- Diamantakos, P.; Ioannidis, K.; Papanikolaou, C.; Tsolakou, A.; Rigakou, A.; Melliou, E.; Magiatis, P. A New Definition of the Term “High-Phenolic Olive Oil” Based on Large Scale Statistical Data of Greek Olive Oils Analyzed by qNMR. Molecules 2021, 26, 1115. [Google Scholar] [CrossRef] [PubMed]
- Grossi, C.; Rigacci, S.; Ambrosini, S.; Dami, T.E.; Luccarini, I.; Traini, C.; Failli, P.; Berti, A.; Casamenti, F.; Stefani, M. The Polyphenol Oleuropein Aglycone Protects TgCRND8 Mice against Aß Plaque Pathology. PLoS ONE 2013, 8, e71702. [Google Scholar]
- Nikou, T.; Liaki, V.; Stathopoulos, P.; Sklirou, A.D.; Tsakiri, E.; Jakschitz, T.; Bonn, G.; Trougakos, I.P.; Halabalaki, M.; Skaltsounis, L.A. Comparison survey of EVOO polyphenols and exploration of healthy aging-promoting properties of oleocanthal and oleacein. Food Chem. Toxicol. 2019, 125, 403–412. [Google Scholar] [CrossRef] [PubMed]
- Kalogeropoulos, N.; Tsimidou, M.Z. Antioxidants in Greek Virgin Olive Oils. Antioxidants 2014, 3, 387–413. [Google Scholar] [CrossRef] [Green Version]
- Klikarová, J.; Rotondo, A.; Cacciola, F.; Česlová, L.; Dugo, P.; Mondello, L.; Rigano, F. The Phenolic Fraction of Italian Extra Virgin Olive Oils: Elucidation Through Combined Liquid Chromatography and NMR Approaches. Food Anal. Methods 2019, 12, 1759–1770. [Google Scholar] [CrossRef]
- García-Rodríguez, R.; Belaj, A.; Romero-Segura, C.; Sanz, C.; Pérez, A.G. Exploration of genetic resources to improve the functional quality of virgin olive oil. J. Funct. Foods 2017, 38, 1–8. [Google Scholar] [CrossRef]
- Agiomyrgianaki, A.; Petrakis, P.V.; Dais, P. Influence of harvest year, cultivar and geographical origin on Greek extra virgin olive oils composition: A study by NMR spectroscopy and biometric analysis. Food Chem. 2012, 135, 2561–2568. [Google Scholar] [CrossRef]
- Diamantakos, P.; Giannara, T.; Skarkou, M.; Melliou, E.; Magiatis, P. Influence of Harvest Time and Malaxation Conditions on the Concentration of Individual Phenols in Extra Virgin Olive Oil Related to Its Healthy Properties. Molecules 2020, 25, 2449. [Google Scholar] [CrossRef] [PubMed]
- Miho, H.; Díez, C.; Mena-Bravo, A.; de Medina, V.S.; Moral, J.; Melliou, E.; Magiatis, P.; Rallo, L.; Barranco, D.; Priego-Capote, F. Cultivar influence on variability in olive oil phenolic profiles determined through an extensive germplasm survey. Food Chem. 2018, 266, 192–199. [Google Scholar] [CrossRef] [PubMed]
- Tsimidou, M.Z. Analytical methodologies: Phenolic compounds related to olive oil taste issues. In Handbook of Olive Oil: Analysis and Properties; Springer: New York, NY, USA, 2013; pp. 311–333. [Google Scholar]
- Miho, H.; Moral, J.; Barranco, D.; Ledesma-Escobar, C.; Priego-Capote, F.; Díez, C. Influence of genetic and interannual factors on the phenolic profiles of virgin olive oils. Food Chem. 2021, 342, 128357. [Google Scholar] [CrossRef]
- Eriotou, E.; Karabagias, I.K.; Maina, S.; Koulougliotis, D.; Kopsahelis, N. Geographical origin discrimination of “Ntopia” olive oil cultivar from Ionian islands using volatile compounds analysis and computational statistics. Eur. Food Res. Technol. 2021, 247, 3083–3098. [Google Scholar] [CrossRef] [PubMed]
- Díez, C.M.; Trujillo, I.; Barrio, E.; Belaj, A.; Barranco, D.; Rallo, L. Centennial olive trees as a reservoir of genetic diversity. Ann. Bot. 2011, 108, 797–807. [Google Scholar] [CrossRef] [Green Version]
- Koulougliotis, D.; Eriotou, E. Isolation and Identification of Endogenous Yeast Strains in Grapes and Must Solids of Mavrodafni kefalonias and Antioxidant Activity of the Produced Red Wine. Ferment. Technol. 2016, 5, 125. [Google Scholar] [CrossRef] [Green Version]
- Eriotou, E.; Anastasiadou, K.; Nikolopoulos, D.; Koulougliotis, D. Antimicrobial and Free Radical Scavenging Activities of Basil (Ocimum basilicum) Essential Oil Isolated from Five Plant Varieties Growing in Greece. J. Nutr. Food Sci. 2014, 5, 1. [Google Scholar] [CrossRef] [Green Version]
- Sika, K.C.; Kefela, T.; Adoukonou-Sagbadja, H.; Ahoton, L.; Saidou, A.; Baba-Moussa, L.; Baptiste, L.J.; Kotconi, S.O.; Gachomo, E.W. A simple and efficient genomic DNA extraction protocol for large scale genetic analyses of plant biological systems. Plant Gene 2015, 1, 43–45. [Google Scholar] [CrossRef] [Green Version]
- Sanz-Cortés, F.; Badenes, M.L.; Paz, S.; Íñiguez, A.; Llácer, G. Molecular Characterization of Olive Cultivars Using RAPD Markers. J. Am. Soc. Hortic. Sci. 2001, 126, 7–12. [Google Scholar] [CrossRef] [Green Version]
- Nei, M. Analysis of Gene Diversity in Subdivided Populations. Proc. Natl. Acad. Sci. USA 1973, 70, 3321–3323. [Google Scholar] [CrossRef] [Green Version]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, R.; Shimodaira, H. Pvclust: An R package for assessing the uncertainty in hierarchical clustering. Bioinformmatics 2006, 22, 1540–1542. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis. In Use R! Springer International Publishing: Cham, Switzerland, 2016. [Google Scholar]
- Cambridge Philosophical Society. Mathematical proceedings of the Cambridge Philosophical Society. In The Testing of Statistical Hypotheses in Relation to Probabilities a Priori; Neyman, J., Pearson, E.S., Eds.; Cambridge Philosophical Society: Cambridge, UK, 2008; pp. 492–510. [Google Scholar]
- Franco, M.N.; Galeano-Díaz, T.; López, Ó.; Fernández-Bolaños, J.G.; Sánchez, J.; De Miguel, C.; Gil, M.V.; Martín-Vertedor, D. Phenolic compounds and antioxidant capacity of virgin olive oil. Food Chem. 2014, 163, 289–298. [Google Scholar] [CrossRef]
- Montaño, A.; Hernández, M.; Garrido, I.; Llerena, J.L.; Espinosa, F. Fatty Acid and Phenolic Compound Concentrations in Eight Different Monovarietal Virgin Olive Oils from Extremadura and the Relationship with Oxidative Stability. Int. J. Mol. Sci. 2016, 17, 1960. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Losito, I.; Abbattista, R.; De Ceglie, C.; Castellaneta, A.; Calvano, C.; Cataldi, T. Bioactive Secoiridoids in Italian Extra-Virgin Olive Oils: Impact of Olive Plant Cultivars, Cultivation Regions and Processing. Molecules 2021, 26, 743. [Google Scholar] [CrossRef] [PubMed]
- Omri, A.; Abdelhamid, S.; Benincasa, C.; Araouki, A.; Ayadi, M.; Gharsallaoui, M.; Gouiaa, M. Genetic diversity and association of molecular markers with biochemical traits in Tunisian olive cultivars. Genet. Resour. Crop. Evol. 2021, 68, 1181–1197. [Google Scholar] [CrossRef]
- Xu, Y.; Liu, X.; Fu, J.; Wang, H.; Wang, J.; Huang, C.; Prasanna, B.M.; Olsen, M.S.; Wang, G.; Zhang, A. Enhancing Genetic Gain through Genomic Selection: From Livestock to Plants. Plant Commun. 2020, 1, 100005. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Niu, E.; Shi, A.; Mou, B. Genetic Diversity Analysis of Olive Germplasm (Olea europaea L.) With Genotyping-by-Sequencing Technology. Front. Genet. 2019, 10, 755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kyriakopoulou, C.; Kalogianni, D.P. Genetic Identification of the Wild Form of Olive (Olea europaea var. sylvestris) Using Allele-Specific Real-Time PCR. Foods 2020, 9, 467. [Google Scholar] [CrossRef] [Green Version]
- D’Agostino, N.; Taranto, F.; Camposeo, S.; Mangini, G.; Fanelli, V.; Gadaleta, S.; Miazzi, M.M.; Pavan, S.; Di Rienzo, V.; Sabetta, W.; et al. GBS-derived SNP catalogue unveiled wide genetic variability and geographical relationships of Italian olive cultivars. Sci. Rep. 2018, 8, 15877. [Google Scholar] [CrossRef] [Green Version]





| Lefkada_Asprolia (n = 35) | Zakynthos_Ntopia (n = 22) | Kefalonia_Ntopia (n = 13) | Kefalonia_Ntopia_old (n = 13) | Kefrkyra_Lianolia (n = 20) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | SD | SEM | Mean | SD | SEM | Mean | SD | SEM | Mean | SD | SEM | Mean | SD | SEM | |
| Antioxidant activity (mmol Trolox/L) | 2.393 | 0.4781 | 0.08081 | 1.893 | 0.4119 | 0.08782 | 1.773 | 0.5961 | 0.1653 | 2.086 | 0.4234 | 0.1174 | 1.994 | 0.3436 | 0.07684 |
| Oleocanthal (mg/Kg) | 39.47 | 27.26 | 4.607 | 176.2 | 65.48 | 13.96 | 181.7 | 64.74 | 17.96 | 261.5 | 49.22 | 13.65 | 273.5 | 81.47 | 18.22 |
| Oleacein (mg/Kg) | 27.04 | 17.6 | 2.975 | 115.2 | 41.6 | 8.869 | 78.42 | 55.15 | 15.3 | 179.5 | 68.19 | 18.91 | 172 | 70.35 | 15.73 |
| Ligstroside aglycon (mg/Kg) | 97.98 | 40.17 | 6.79 | 35.08 | 21.06 | 4.49 | 25.78 | 16.62 | 4.61 | 35.27 | 9.708 | 2.692 | 19.19 | 15.99 | 3.575 |
| Oleuropein aglycon (mg/Kg) | 178 | 78.82 | 13.32 | 64.38 | 66.73 | 14.23 | 26.49 | 22.58 | 6.263 | 38.85 | 18.02 | 4.997 | 6.627 | 10.41 | 2.327 |
| Oleokoronal (mg/Kg) | 90.56 | 42.91 | 7.254 | 41.65 | 21.13 | 4.506 | 19.34 | 33.03 | 9.16 | 50.12 | 47.83 | 13.27 | 14.92 | 13.03 | 2.913 |
| Oleomissional (mg/Kg) | 102.5 | 61.33 | 10.37 | 37.77 | 37.62 | 8.021 | 15.06 | 30.14 | 8.36 | 20.87 | 25.8 | 7.155 | 4.57 | 0 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kalaboki, I.; Koulougliotis, D.; Kleisiari, D.; Melliou, E.; Magiatis, P.; Kampioti, A.; Eriotou, E.; Destouni, A. A Study on the Clustering of Extra Virgin Olive Oils Extracted from Cultivars Growing in Four Ionian Islands (Greece) by Multivariate Analysis of Their Phenolic Profile, Antioxidant Activity and Genetic Markers. Foods 2021, 10, 3009. https://0-doi-org.brum.beds.ac.uk/10.3390/foods10123009
Kalaboki I, Koulougliotis D, Kleisiari D, Melliou E, Magiatis P, Kampioti A, Eriotou E, Destouni A. A Study on the Clustering of Extra Virgin Olive Oils Extracted from Cultivars Growing in Four Ionian Islands (Greece) by Multivariate Analysis of Their Phenolic Profile, Antioxidant Activity and Genetic Markers. Foods. 2021; 10(12):3009. https://0-doi-org.brum.beds.ac.uk/10.3390/foods10123009
Chicago/Turabian StyleKalaboki, Iliana, Dionysios Koulougliotis, Dimitra Kleisiari, Eleni Melliou, Prokopios Magiatis, Adamantia Kampioti, Effimia Eriotou, and Aspasia Destouni. 2021. "A Study on the Clustering of Extra Virgin Olive Oils Extracted from Cultivars Growing in Four Ionian Islands (Greece) by Multivariate Analysis of Their Phenolic Profile, Antioxidant Activity and Genetic Markers" Foods 10, no. 12: 3009. https://0-doi-org.brum.beds.ac.uk/10.3390/foods10123009