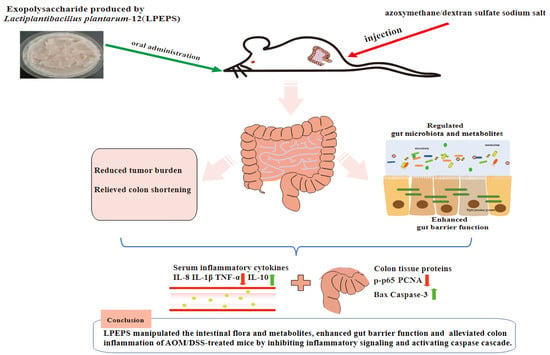
Exopolysaccharide Produced by Lactiplantibacillus plantarum-12 Alleviates Intestinal Inflammation and Colon Cancer Symptoms by Modulating the Gut Microbiome and Metabolites of C57BL/6 Mice Treated by Azoxymethane/Dextran Sulfate Sodium Salt
Abstract
:1. Introduction
2. Materials and Methods
2.1. Material and Reagents
2.2. Preparation of the LPEPS
2.3. Animal Experiment Design
2.4. Histological and Biochemical Analysis
2.5. Western Blot Analysis
2.6. Gut Microbiota Analysis
2.7. Fecal Metabolomic Analysis
2.8. Statistical Analysis
3. Results
3.1. Effect of LPEPS Oral Administration on the Body Weight, Colon Length, and Tumor Numbers of the AOM/DSS-Treated C57BL/6 Mice
3.2. Effect of LPEPS Oral Administration on Serum Inflammatory Cytokine and Organ Index of the AOM/DSS-Treated C57BL/6 Mice
3.3. Effect of LPEPS Oral Administration on Gut Barrier Function of the AOM/DSS-Treated C57BL/6 Mice
3.4. Effect of LPEPS Oral Administration on the Expression of Colonic Inflammation-Related Proteins of the AOM/DSS-Treated C57BL/6 Mice
3.5. Effect of LPEPS Oral Administration on the Expression of Colonic Apoptosis-Related Proteins of the AOM/DSS-Treated C57BL/6 Mice
3.6. Effect of LPEPS Oral Administration on the Intestinal Flora of the AOM/DSS-Treated C57BL/6 Mice
3.7. Effect of LPEPS Oral Administration on the Fecal Metabolites of the AOM/DSS-Treated C57BL/6 Mice
3.8. Correlation Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Keefe, S.J.D. Diet, microorganisms and their metabolites, and colon cancer. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 691–706. [Google Scholar] [CrossRef] [PubMed]
- Louis, P.; Hold, G.L.; Flint, H.J. The gut microbiota, bacterial metabolites and colorectal cancer. Nat. Rev. Microbiol. 2014, 12, 661–672. [Google Scholar] [CrossRef] [PubMed]
- Clos-Garcia, M.; Garcia, K.; Alonso, C.; Iruarrizaga-Lejarreta, M.; D’Amato, M.; Crespo, A.; Iglesias, A.; Cubiella, J.; Bujanda, L.; Falcón-Pérez, J.M. Integrative analysis of fecal metagenomics and metabolomics in colorectal cancer. Cancers 2020, 12, 1142. [Google Scholar] [CrossRef]
- Jackson, L.N.; Zhou, Y.; Qiu, S.; Wang, Q.; Evers, B.M. Alternative Medicine Products as a Novel Treatment Strategy for Inflammatory Bowel Disease. Am. J. Chin. Med. 2008, 36, 953–965. [Google Scholar] [CrossRef] [PubMed]
- Nadeem, M.S.; Kumar, V.; Al-Abbasi, F.A.; Kamal, M.A.; Anwar, F. Risk of colorectal cancer in inflammatory bowel diseases. Semin. Cancer Biol. 2020, 64, 51–60. [Google Scholar] [CrossRef]
- Wu, J.; Zhang, Y.; Ye, L.; Wang, C. The anti-cancer effects and mechanisms of lactic acid bacteria exopolysaccharides in vitro: A review. Carbohydr. Polym. 2021, 253, 117308. [Google Scholar] [CrossRef] [PubMed]
- Ji, X.; Hou, C.; Gao, Y.; Xue, Y.; Yan, Y.; Guo, X. Metagenomic analysis of gut microbiota modulatory effects of jujube (Ziziphus jujuba Mill.) polysaccharides in a colorectal cancer mouse model. Food Funct. 2020, 11, 163–173. [Google Scholar] [CrossRef]
- Smith, B.J.; Miller, R.A.; Ericsson, A.C.; Harrison, D.C.; Strong, R.; Schmidt, T.M. Changes in the gut microbiome and fermentation products concurrent with enhanced longevity in acarbose-treated mice. BMC Microbiol. 2019, 19, 130. [Google Scholar] [CrossRef] [Green Version]
- Tian, B.; Zhao, J.; Zhang, M.; Chen, Z.; Ma, Q.; Liu, H.; Nie, C.; Zhang, Z.; An, W. Lycium ruthenicum Anthocyanins Attenuate High-Fat Diet-Induced Colonic Barrier Dysfunction and Inflammation in Mice by Modulating the Gut Microbiota. Mol. Nutr. Food Res. 2021, 65, 2000745. [Google Scholar] [CrossRef]
- Damazo-Lima, M.; Rosas-Pérez, G.; Reynoso-Camacho, R.; Pérez-Ramírez, I.F.; Rocha-Guzmán, N.E.; De Los Ríos, E.A.; Ramos-Gomez, M. Chemopreventive Effect of the Germinated Oat and Its Phenolic-AVA Extract in Azoxymethane/Dextran Sulfate Sodium (AOM/DSS) Model of Colon Carcinogenesis in Mice. Foods 2020, 9, 169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Qin, J.; Cheng, Y.; Ai, Y.; Han, Z.; Li, M.; Qi, Y.; Zhao, Q.; Li, Z. Polysaccharide from Patinopecten yessoensis Skirt Boosts Immune Response via Modulation of Gut Microbiota and Short-Chain Fatty Acids Metabolism in Mice. Foods 2021, 10, 2478. [Google Scholar] [CrossRef]
- Luo, J.; Zhang, C.; Liu, R.; Gao, L.; Ou, S.; Liu, L.; Peng, X. Ganoderma lucidum polysaccharide alleviating colorectal cancer by alteration of special gut bacteria and regulation of gene expression of colonic epithelial cells. J. Funct. Foods 2018, 47, 127–135. [Google Scholar] [CrossRef]
- Zahran, W.E.; Elsonbaty, S.M.; Moawed, S.M.F. Lactobacillus rhamnosus ATCC 7469 exopolysaccharides synergizes with low level ionizing radiation to modulate signaling molecular targets in colorectal carcinogenesis in rats. Biomed. Pharmacother. 2017, 92, 384–393. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Wang, J.; Ji, Y.; Chen, K. Metabonomic Variation of Exopolysaccharide from Rhizopus Nigricans on AOM/DSS-Induced Colorectal Cancer in Mice. OncoTargets Ther. 2019, 12, 10023–10033. [Google Scholar] [CrossRef] [Green Version]
- Matsuzaki, C.; Hayakawa, A.; Matsumoto, K.; Katoh, T.; Yamamoto, K.; Hisa, K. Exopolysaccharides Produced by Leuconostoc mesenteroides Strain NTM048 as an Immunostimulant to Enhance the Mucosal Barrier and Influence the Systemic Immune Response. J. Agric. Food Chem. 2015, 63, 7009–7015. [Google Scholar] [CrossRef]
- London, L.E.; Kumar, A.H.; Wall, R.; Casey, P.G.; O’ Sullivan, O.; Shanahan, F.; Hill, C.; Cotter, P.D.; Fitzgerald, G.F.; Ross, R.P.; et al. Exopolysaccharide-Producing Probiotic Lactobacilli Reduce Serum Cholesterol and Modify Enteric Microbiota in ApoE-Deficient Mice. J. Nutr. 2014, 144, 1956–1962. [Google Scholar] [CrossRef] [Green Version]
- Sun, M.; Liu, W.; Song, Y.; Tuo, Y.; Mu, G.; Ma, F. The Effects of Lactobacillus plantarum-12 Crude Exopolysaccharides on the Cell Proliferation and Apoptosis of Human Colon Cancer (HT-29) Cells. Probiotics Antimicrob. Proteins 2020, 20, 413–421. [Google Scholar] [CrossRef]
- Song, Y.; Sun, M.; Feng, L.; Liang, X.; Song, X.; Mu, G.; Tuo, Y.; Jiang, S.; Qian, F. Lactobacillus plantarum-12 Exopolysaccharides Have Anti-Biofilm Activity Against Shigella flexneri. Appl. Environ. Microbiol. 2020, 86, e00694-20. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Gao, Y.; Ma, F.; Sun, M.; Mu, G.; Tuo, Y. The ameliorative effect of Lactobacillus plantarum Y44 oral administration on inflammation and lipid metabolism in obese mice fed with a high fat diet. Food Funct. 2020, 11, 5024–5039. [Google Scholar] [CrossRef]
- Wang, X.; Sun, G.; Feng, T.; Zhang, J.; Huang, X.; Wang, T.; Xie, Z.; Chu, X.; Yang, J.; Wang, H.; et al. Sodium oligomannate therapeutically remodels gut microbiota and suppresses gut bacterial amino acids-shaped neuroinflammation to inhibit Alzheimer’s disease progression. Cell Res. 2019, 29, 787–803. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.; Zeng, B.; Hu, L.; Zhang, Y.; Xiong, J.; Deng, J.; Huang, L.; Liao, Z.; Wang, J.; Wei, H.; et al. Effect of a Humanized Diet Profile on Colonization Efficiency and Gut Microbial Diversity in Human Flora-Associated Mice. Front. Nutr. 2021, 8, 676085. [Google Scholar] [CrossRef]
- Borges-Canha, M.; Portela-Cidade, J.P.; Dinis-Ribeiro, M.; Leite-Moreira, A.F.; Pimentel-Nunes, P. Role of colonic microbiota in colorectal carcinogenesis: A systematic review. Rev. Esp. Enferm. Dig. 2015, 107, 659–671. [Google Scholar] [CrossRef] [Green Version]
- Mendes, M.C.S.; Paulino, D.S.M.; Brambilla, S.R.; Camargo, J.A.; Persinoti, G.F.; Carvalheira, J.B.C. Microbiota modification by probiotic supplementation reduces colitis associated colon cancer in mice. World J. Gastroenterol. 2018, 24, 1995–2008. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Niu, Z.; Zou, M.; Liu, S.; Wang, M.; Gu, X.; Lu, H.; Tian, H.; Jha, R. Probiotics, prebiotics, and synbiotics regulate the intestinal microbiota differentially and restore the relative abundance of specific gut microorganisms. J. Dairy Sci. 2020, 103, 5816–5829. [Google Scholar] [CrossRef]
- Lagkouvardos, I.; Lesker, T.R.; Hitch, T.C.A.; Gálvez, E.J.C.; Smit, N.; Neuhaus, K.; Wang, J.; Baines, J.F.; Abt, B.; Stecher, B.; et al. Sequence and cultivation study of Muribaculaceae reveals novel species, host preference, and functional potential of this yet undescribed family. Microbiome 2019, 7, 28. [Google Scholar] [CrossRef] [Green Version]
- Smith, B.J.; Miller, R.A.; Schmidt, T.M. Muribaculaceae genomes assembled from metagenomes suggest genetic drivers of differential response to acarbose treatment in mice. mSphere 2021, e0085121. [Google Scholar] [CrossRef]
- Beerens, H.; Romond, C. Sulfate-reducing anaerobic bacteria in human feces. Am. J. Clin. Nutr. 1977, 30, 1770–1776. [Google Scholar] [CrossRef] [Green Version]
- Zhai, Z.; Zhang, F.; Cao, R.; Ni, X.; Xin, Z.; Deng, J.; Wu, G.; Ren, W.; Yin, Y.; Deng, B. Cecropin A Alleviates Inflammation Through Modulating the Gut Microbiota of C57BL/6 Mice With DSS-Induced IBD. Front. Microbiol. 2019, 10, 1595. [Google Scholar] [CrossRef]
- Zhao, Y.; Liu, S.; Tang, Y.; You, T.; Xu, H. Lactobacillus rhamnosus GG Ameliorated Long-Term Exposure to TiO2 Nanoparticles Induced Microbiota-Mediated Liver and Colon Inflammation and Fructose-Caused Metabolic Abnormality in Metabolism Syndrome Mice. J. Agric. Food Chem. 2021, 69, 9788–9799. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Wilson, J.E.; Koenigsknecht, M.J.; Chou, W.C.; Montgomery, S.A.; Truax, A.D.; Brickey, W.J.; Packey, C.D.; Maharshak, N.; Matsushima, G.K.; et al. NLRP12 attenuates colon inflammation by maintaining colonic microbial diversity and promoting protective commensal bacterial growth. Nat. Immunol. 2017, 18, 541–551. [Google Scholar] [CrossRef] [PubMed]
- Taddese, R.; Garza, D.R.; Ruiter, L.N.; De Jonge, M.I.; Belzer, C.; Aalvink, S.; Nagtegaal, I.D.; Dutilh, B.E.; Boleij, A. Growth rate alterations of human colorectal cancer cells by 157 gut bacteria. Gut Microbes 2020, 12, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Saito, K.; Koido, S.; Odamaki, T.; Kajihara, M.; Kato, K.; Horiuchi, S.; Adachi, S.; Arakawa, H.; Yoshida, S.; Akasu, T.; et al. Metagenomic analyses of the gut microbiota associated with colorectal adenoma. PLoS ONE 2019, 14, e0212406. [Google Scholar] [CrossRef]
- Koliarakis, I.; Psaroulaki, A.; Nikolouzakis, T.K.; Kokkinakis, M.; Sgantzos, M.; Goulielmos, G.; Androutsopoulos, V.P.; Tsatsakis, A.; Tsiaoussis, J. Intestinal microbiota and colorectal cancer: A new aspect of research. Off. J. Balk. Union Oncol. 2018, 23, 1216–1234. [Google Scholar]
- Kaakoush, N.O. Insights into the Role of Erysipelotrichaceae in the Human Host. Front. Cell. Infect. Microbiol. 2015, 5, 84. [Google Scholar] [CrossRef] [Green Version]
- Yan, S.; Yang, B.; Zhao, J.; Zhao, J.; Stanton, C.; Ross, R.P.; Zhang, H.; Chen, W. A ropy exopolysaccharide producing strain Bifidobacterium longum subsp. longum YS108R alleviates DSS-induced colitis by maintenance of the mucosal barrier and gut microbiota modulation. Food Funct. 2019, 10, 1595–1608. [Google Scholar] [CrossRef]
- Scarpa, M.; Grillo, A.; Faggian, D.; Ruffolo, C.; Bonello, E.; D’Incà, R.; Scarpa, M.; Castagliuolo, I.; Angriman, I. Relationship between mucosa-associated microbiota and inflammatory parameters in the ileal pouch after restorative proctocolectomy for ulcerative colitis. Surgery 2011, 150, 56–67. [Google Scholar] [CrossRef]
- Tannock, G.W.; Lawley, B.; Munro, K.; Lay, C.; Taylor, C.; Daynes, C.; Baladjay, L.; Mcleod, R.; Thompson-Fawcett, M. Comprehensive analysis of the bacterial content of stool from patients with chronic pouchitis, normal pouches, or familial adenomatous polyposis pouches. Inflamm. Bowel Dis. 2012, 18, 925–934. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Yin, W.; Zhang, Y.; Lv, Q.; Yang, Y.; He, J. Foes or Friends? Bacteria Enriched in the Tumor Microenvironment of Colorectal Cancer. Cancers 2020, 12, 372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Tang, L.; Zhou, H.; Zhou, J.; Glenn, T.C.; Shen, C.L.; Wang, J.S. Long-term treatment with green tea polyphenols modifies the gut microbiome of female sprague-dawley rats. J. Nutr. Biochem. 2018, 56, 55–64. [Google Scholar] [CrossRef]
- Zhou, X.; Zhang, D.; Qi, W.; Hong, T.; Wu, T.; Geng, F.; Geng, F.; Xie, M.; Nie, S. Exopolysaccharides from Lactobacillus plantarum NCU116 Facilitate Intestinal Homeostasis by Modulating Intestinal Epithelial Regeneration and Microbiota. J. Agric. Food Chem. 2017, 7, 7863–7873. [Google Scholar] [CrossRef]
- Bryan, G.T. The role of urinary tryptophan metabolites in the etiology of bladder cancer. Am. J. Clin. Nutr. 1971, 24, 841–847. [Google Scholar] [CrossRef] [PubMed]
- Wan, Y.; Wang, F.; Yuan, J.; Li, J.; Jiang, D.; Zhang, J.; Li, H.; Wang, R.; Tang, J.; Huang, T.; et al. Effects of dietary fat on gut microbiota and faecal metabolites, and their relationship with cardiometabolic risk factors: A 6-month randomised controlled-feeding trial. Gut 2019, 68, 1417–1429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barreto, F.C.; Barreto, D.V.; Liabeuf, S.; Meert, N.; Glorieux, G.; Temmar, M.; Choukroun, G.; Vanholder, R.; Massy, Z.A. Serum Indoxyl Sulfate Is Associated with Vascular Disease and Mortality in Chronic Kidney Disease Patients. Clin. J. Am. Soc. Nephrol. 2009, 4, 1551–1558. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Zhao, F.; Wang, T.; Xu, Y.; Qiu, J.; Qian, Y. Host Metabolic Disorders Induced by Alterations in Intestinal Flora under Dietary Pesticide Exposure. J. Agric. Food Chem. 2021, 69, 6303–6317. [Google Scholar] [CrossRef]
- Nie, Q.; Xing, M.; Chen, H.; Hu, J.; Nie, S. Metabolomics and Lipidomics Profiling Reveals Hypocholesterolemic and Hypolipidemic Effects of Arabinoxylan on Type 2 Diabetic Rats. J. Agric. Food Chem. 2019, 67, 10614–10623. [Google Scholar] [CrossRef]
- Zhao, L.; Zhang, F.; Ding, X.; Wu, G.; Lam, Y.Y.; Wang, X.; Fu, H.; Xue, X.; Lu, C.; Zhang, C.; et al. Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science 2018, 359, 1151–1156. [Google Scholar] [CrossRef] [Green Version]
- Ding, H.; Zhao, X.; Ma, C.; Gao, Q.; Yin, Y.; Kong, X.; He, J. Dietary Supplementation with Bacillus subtilis DSM 32315 Alters the Intestinal Microbiota and Metabolites in Weaned Piglets. J. Appl. Microbiol. 2020, 130, 217–232. [Google Scholar] [CrossRef]
- Ballongue, J.; Schumann, C.; Quignon, P. Effects of Lactulose and Lactitol on Colonic Microflora and Enzymatic Activity. Scand. J. Gastroenterol. 1997, 32, 41–44. [Google Scholar] [CrossRef]
- Nie, Q.; Hu, J.; Gao, H.; Li, M.; Sun, Y.; Chen, H.; Zuo, S.; Fang, Q.; Huang, X.; Yin, J.; et al. Bioactive Dietary Fibers Selectively Promote Gut Microbiota to Exert Antidiabetic Effects. J. Agric. Food Chem. 2021, 69, 7000–7015. [Google Scholar] [CrossRef] [PubMed]
- Hsu, W.Y.; Chen, W.T.; Lin, W.D.; Tsai, F.J.; Tsai, Y.; Lin, C.T.; Lo, W.Y.; Jeng, L.B.; Lai, C.C. Analysis of urinary nucleosides as potential tumor markers in human colorectal cancer by high performance liquid chromatography/electrospray ionization tandem mass spectrometry. Clin. Chim. Acta. 2009, 402, 31–37. [Google Scholar] [CrossRef]
- Garza, D.R.; Taddese, R.; Wirbel, J.; Zeller, G.; Boleij, A.; Huynen, M.A.; Dutilh, B.E. Metabolic models predict bacterial passengers in colorectal cancer. Cancer Metab. 2020, 8, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, E.C.Y.; Koh, P.K.; Mal, M.; Cheah, P.Y.; Eu, K.W.; Backshall, A.; Cavill, R.; Nicholson, J.K.; Keun, H.C. Metabolic Profiling of Human Colorectal Cancer Using High-Resolution Magic Angle Spinning Nuclear Magnetic Resonance (HR-MAS NMR) Spectroscopy and Gas Chromatography Mass Spectrometry (GC/MS). J. Proteome Res. 2009, 8, 352–361. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Gowda, G.A.N.; Zhu, J.; Deng, L.; Gu, H.; Chiorean, E.G.; Zaid, M.A.; Harrison, M.; Zhang, D.; Zhang, M.; et al. Altered metabolite levels and correlations in patients with colorectal cancer and polyps detected using seemingly unrelated regression analysis. Metabolomics 2017, 13, 125. [Google Scholar] [CrossRef]
- Brown, D.G.; Rao, S.; Weir, T.L.; O’Malia, J.; Bazan, M.; Brown, R.J.; Ryan, E.P. Metabolomics and metabolic pathway networks from human colorectal cancers, adjacent mucosa, and stool. Cancer Metab. 2016, 4, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, C.H.; Dejea, C.M.; Edler, D.; Hoang, L.T.; Santidrian, A.F.; Felding, B.H.; Ivanisevic, J.; Cho, K.; Wick, E.C.; Hechenbleikner, E.M.; et al. Metabolism Links Bacterial Biofilms and Colon Carcinogenesis. Cell Metab. 2015, 21, 891–897. [Google Scholar] [CrossRef] [Green Version]
- Williams, M.D.; Zhang, X.; Park, J.-J.; Siems, W.F.; Gang, D.R.; Resar, L.M.S.; Reeves, R.; Hill Jr, H.H. Characterizing metabolic changes in human colorectal cancer. Anal. Bioanal. Chem. 2015, 407, 4581–4595. [Google Scholar] [CrossRef] [PubMed]
- Gumpenberger, T.; Brezina, S.; Keski-Rahkonen, P.; Baierl, A.; Robinot, N.; Leeb, G.; Habermann, N.; Kok, D.E.G.; Scalbert, A.; Ueland, P.-M.; et al. Untargeted metabolomics reveals major differences in the plasma metabolome between colorectal cancer and colorectal adenomas. Metabolites 2021, 11, 119. [Google Scholar] [CrossRef]
- Zong, L.; Xing, J.; Liua, S.; Liu, Z.; Song, F. Cell metabolomics reveals the neurotoxicity mechanism of cadmium in PC12 cells. Ecotoxicol. Environ. Saf. 2017, 147, 16–33. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, B.; Huang, S.; Wang, F.; Zheng, L.; Lu, J.; Zeng, Y.; Chen, J.; Li, S. Metabolomics Analysis Reveals the Protection Mechanism of Huangqi–Danshen Decoction on Adenine-Induced Chronic Kidney Disease in Rats. Front. Pharmacol. 2019, 10, 992. [Google Scholar] [CrossRef]
- Pantophlet, A.J.; Roelofsen, H.; De Vries, M.P.; Gerrits, W.J.J.; Van Den Borne, J.J.G.C.; Vonk, R.J. The use of metabolic profiling to identify insulin resistance in veal calves. PLoS ONE 2017, 12, e0179612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Birt, D.F.; Hendrich, S.; Wang, W. Dietary agents in cancer prevention: Flavonoids and isoflavonoids. Pharmacol. Ther. 2001, 90, 157–177. [Google Scholar] [CrossRef]
- Tang, D.; Shen, Y.-B.; Wang, Z.-H.; He, B.; Xu, Y.-H.; Nie, H.; Zhu, Q. Rapid Analysis and Guided Isolation of Astragalus Isoflavonoids by UHPLC-DAD-MSn and Their Cellular Antioxidant Defense on High Glucose Induced Mesangial Cells Dysfunction. J. Agric. Food Chem. 2018, 66, 1105–1113. [Google Scholar] [CrossRef] [PubMed]
- Hämäläinen, M.; Nieminen, R.; Vuorela, P.; Heinonen, M.; Moilanen, E. Anti-Inflammatory Effects of Flavonoids: Genistein, Kaempferol, Quercetin, and Daidzein Inhibit STAT-1 and NF-κB Activations, Whereas Flavone, Isorhamnetin, Naringenin, and Pelargonidin Inhibit only NF-κB Activation along with Their Inhibitory Effect on iNOS Expression and NO Production in Activated Macrophages. Mediat. Inflamm. 2007, 2007, 045673. [Google Scholar] [CrossRef] [Green Version]
- Phang, J.M.; Liu, W.; Zabirnyk, O. Proline Metabolism and Microenvironmental Stress. Annu. Rev. Nutr. 2010, 30, 441–463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, G.; Bazer, F.W.; Burghardt, R.C.; Johnson, G.A.; Kim, S.W.; Knabe, D.A.; Li, P.; Li, X.; McKnight, J.R.; Satterfield, M.C.; et al. Proline and hydroxyproline metabolism: Implications for animal and human nutrition. Amino Acids 2010, 40, 1053–1063. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, F.; Kim, S.; Yin, X.; Zhang, X.; Kato, I. Integrating Two-Dimensional Gas and Liquid Chromatography-Mass Spectrometry for Untargeted Colorectal Cancer Metabolomics: A Proof-of-Principle Study. Metabolites 2020, 10, 343. [Google Scholar] [CrossRef] [PubMed]
- Geijsen, A.; Brezina, S.; Keski-Rahkonen, P.; Baierl, A.; Bachleitner-Hofmann, T.; Bergmann, M.M.; Boehm, J.; Brenner, H.; Chang-Claude, J.; van Duijnhoven, F.; et al. Plasma metabolites associated with colorectal cancer: A discovery-replication strategy. Int. J. Cancer 2019, 145, 1221–1231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, M.; Zhang, X.; Wang, Y.; Li, Y.; Chen, Y.; Zheng, H.; Ma, F.; Ma, C.W.; Lu, B.; Xie, Z.; et al. Metabonomic strategy for the detection of metabolic effects of probiotics combined with prebiotic supplementation in weaned rats. RSC Adv. 2018, 8, 5042–5057. [Google Scholar] [CrossRef] [Green Version]
- Dorrestein, P.C.; Mazmanian, S.K.; Knight, R. Finding the Missing Links among Metabolites, Microbes, and the Host. Immunity 2014, 40, 824–832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, L.; Yang, M.; Dong, W.; Liu, T.; Song, X.; Gu, Y.; Wang, S.; Liu, Y.; Abla, Z.; Qiao, X.; et al. Gut Dysbiosis and Abnormal Bile Acid Metabolism in Colitis-Associated Cancer. Gastroenterology 2021, 2021, 6645970. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.Q.; Nie, S.P.; Shen, M.Y.; Hu, J.L.; Yu, Q.; Gong, D.; Xie, M.Y. Tea Polysaccharides Inhibit Colitis-Associated Colorectal Cancer via Interleukin-6/STAT3 Pathway. J. Agric. Food Chem. 2018, 66, 4384–4393. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.; Yan, T.; Tong, Y.; Deng, H.; Tan, C.; Wan, M.; Wang, M.; Meng, X.; Wang, Y. Gut Microbiota Modulation by Polyphenols from Aronia melanocarpa of LPS-Induced Liver Diseases in Rats. J. Agric. Food Chem. 2021, 69, 3312–3325. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Qi, W.; Hong, T.; Xiong, T.; Gong, D.; Xie, M.; Nie, S. Exopolysaccharides from Lactobacillus plantarum NCU116 regulate intestinal epithelial barrier function via STAT3 signaling pathway. J. Agric. Food Chem. 2018, 66, 9719–9727. [Google Scholar] [CrossRef]
- Sun, M.; Liu, Y.; Song, Y.; Gao, Y.; Zhao, F.; Luo, Y.; Qian, F.; Mu, G.; Tuo, Y. The ameliorative effect of Lactobacillus plantarum-12 on DSS-induced murine colitis. Food Funct. 2020, 11, 5205–5222. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Ren, D.; Yan Zhao, L.L.; Yang, X. Fuzhuan Brick Tea Polysaccharide Improved Ulcerative Colitis in Association with Gut Microbiota-Derived Tryptophan Metabolism. J. Agric. Food Chem. 2021, 69, 8448–8459. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Sun, J.; Liu, J.; Gou, Y.; Zhang, X.; Wu, X.; Sun, R.; Tang, S.; Kan, J.; Qian, C.; et al. Structural characterization and anti-inflammatory activity of alkali-soluble polysaccharides from purple sweet potato Hong. Int. J. Biol. Macromol. 2019, 131, 484–494. [Google Scholar] [CrossRef]
- Li, L.; Jiang, Y.-J.; Yang, X.-Y.; Liu, Y.; Wang, J.-Y.; Man, C.-X. Immunoregulatory effects on Caco-2 cells and mice of exopolysaccharides isolated from Lactobacillus acidophilus NCFM. Food Funct. 2014, 5, 3261–3268. [Google Scholar] [CrossRef]
- Song, G.; Lu, Y.; Yu, Z.; Xu, L.; Liu, J.; Chen, K.; Zhang, P. The inhibitory effect of polysaccharide from Rhizopus nigricans on colitis-associated colorectal cancer. Biomed. Pharmacother. 2019, 112, 108593. [Google Scholar] [CrossRef] [PubMed]
- Chung, K.S.; Cheon, S.Y.; Roh, S.S.; Lee, M.; An, H.J. Chemopreventive Effect of Aster glehni on Inflammation-Induced Colorectal Carcinogenesis in Mice. Nutrients 2018, 10, 202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ben-Neriah, Y.; Karin, M. Inflammation meets cancer, with NF-κB as the matchmaker. Nat. Immunol. 2011, 12, 715–723. [Google Scholar] [CrossRef] [PubMed]
- Pikarsky, E.; Porat, R.M.; Stein, I.; Abramovitch, R.; Amit, S.; Kasem, S.; Gutkovich-Pyest, E.; Urieli-Shoval, S.; Galun, E.; Ben-Neriah, Y. NF-kappaB functions as a tumour promoter in inflammation-associated cancer. Nature 2004, 431, 461–466. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.; Karin, M. Mammalian, MAP kinase signalling cascades. Nature 2001, 410, 37–40. [Google Scholar] [CrossRef]
- Herrington, F.D.; Carmody, R.J.; Goodyear, C.S. Modulation of NF-κB Signaling as a Therapeutic Target in Autoimmunity. J. Biomol. Screen. 2016, 21, 223–242. [Google Scholar] [CrossRef] [Green Version]
- Peng, C.; Ouyang, Y.; Lu, N.; Li, N. The NF-κB Signaling Pathway, the Microbiota, and Gastrointestinal Tumorigenesis: Recent Advances. Front. Immunol. 2020, 11, 1387. [Google Scholar] [CrossRef] [PubMed]
- Bagheri, E.; Hajiaghaalipour, F.; Nyamathulla, S.; Salehen, N.A. The apoptotic effects of Brucea javanica fruit extract against hT29 cells associated with p53 upregulation and inhibition of nF-κB translocation. Drug Des. Dev. Ther. 2018, 12, 657–671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, G.; Qian, W.; Li, J.; Xu, Y.; Chen, K. Exopolysaccharide of Antarctic bacterium Pseudoaltermonas sp. S-5 induces apoptosis in K562 cells. Carbohydr. Polym. 2015, 121, 107–114. [Google Scholar] [CrossRef] [PubMed]

















Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, F.; Song, Y.; Sun, M.; Wang, A.; Jiang, S.; Mu, G.; Tuo, Y. Exopolysaccharide Produced by Lactiplantibacillus plantarum-12 Alleviates Intestinal Inflammation and Colon Cancer Symptoms by Modulating the Gut Microbiome and Metabolites of C57BL/6 Mice Treated by Azoxymethane/Dextran Sulfate Sodium Salt. Foods 2021, 10, 3060. https://0-doi-org.brum.beds.ac.uk/10.3390/foods10123060
Ma F, Song Y, Sun M, Wang A, Jiang S, Mu G, Tuo Y. Exopolysaccharide Produced by Lactiplantibacillus plantarum-12 Alleviates Intestinal Inflammation and Colon Cancer Symptoms by Modulating the Gut Microbiome and Metabolites of C57BL/6 Mice Treated by Azoxymethane/Dextran Sulfate Sodium Salt. Foods. 2021; 10(12):3060. https://0-doi-org.brum.beds.ac.uk/10.3390/foods10123060
Chicago/Turabian StyleMa, Fenglian, Yinglong Song, Mengying Sun, Arong Wang, Shujuan Jiang, Guangqing Mu, and Yanfeng Tuo. 2021. "Exopolysaccharide Produced by Lactiplantibacillus plantarum-12 Alleviates Intestinal Inflammation and Colon Cancer Symptoms by Modulating the Gut Microbiome and Metabolites of C57BL/6 Mice Treated by Azoxymethane/Dextran Sulfate Sodium Salt" Foods 10, no. 12: 3060. https://0-doi-org.brum.beds.ac.uk/10.3390/foods10123060