A Novel Fluorescent Aptasensor Based on Real-Time Fluorescence and Strand Displacement Amplification for the Detection of Ochratoxin A
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents and Apparatus
2.2. OTA Detection by the Fluorescent Aptasensor
2.3. Specificity and Reproducibility Analysis
2.4. Polyacrylamide Gel Electrophoresis (PAGE) Analysis
2.5. Preparation of Real Samples and Detection
3. Results and Discussion
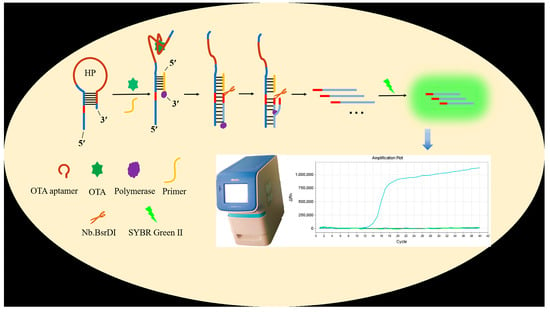
3.1. Principle of Fluorescent Aptasensor
3.2. The Feasibility of OTA Fluorescent Aptasensor
3.3. Optimization of the Experimental Conditions
3.4. Analytical Performance
3.5. Specificity and Reproducibility
3.6. Real Samples Detection
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Chen, R.; Sun, Y.; Huo, B.; Zhao, X.; Huang, H.; Li, S.; Bai, J.; Liang, J.; Gao, Z. A copper monosulfide-nanoparticle-based fluorescent probe for the sensitive and specific detection of ochratoxin A. Talanta 2021, 222, 121678. [Google Scholar] [CrossRef] [PubMed]
- Taghdisi, S.M.; Danesh, N.M.; Ramezani, M.; Alibolandi, M.; Nameghi, M.A.; Gerayelou, G.; Abnous, K. A novel electrochemical aptasensor for ochratoxin a sensing in spiked food using strand-displacement polymerase reaction. Talanta 2021, 223, 121705. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Chen, Y.; Wu, Y.; Weng, B.; Liu, Y.; Lu, Z.; Li, C.M.; Yu, C. Aptamer induced assembly of fluorescent nitrogen-doped carbon dots on gold nanoparticles for sensitive detection of AFB1. Biosens. Bioelectron. 2016, 78, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Han, B.; Fang, C.; Sha, L.; Jalalah, M.; Al-Assiri, M.S.; Harraz, F.A.; Cao, Y. Cascade strand displacement reaction-assisted aptamer-based highly sensitive detection of ochratoxin A. Food Chem. 2021, 338, 127827. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Huang, Q.; Tanveer, Z.I.; Jiang, K.; Zhang, J.; Pan, H.; Luan, L.; Liu, X.; Han, Z.; Wu, Y. “Turn off-on” fluorescent sensor based on quantum dots and self-assembled porphyrin for rapid detection of ochratoxin A. Sens. Actuator B-Chem. 2020, 302, 127212. [Google Scholar] [CrossRef]
- Vidal, A.; Marín, S.; Morales, H.; Ramos, A.J.; Sanchis, V. The fate of deox-ynivalenol and ochratoxin A during the breadmaking process, effects of sourdough use and bran content. Food Chem. Toxicol. 2014, 68, 53–60. [Google Scholar] [CrossRef]
- Zuckerman, A.J. IARC Monographs on the Evaluation of Carcinogenic Risks to Humans. J. Clin. Pathol. 1995, 48, 691. [Google Scholar]
- Badie Bostan, H.; Danesh, N.M.; Karimi, G.; Ramezani, M.; Mousavi Shaegh, S.A.; Youssefi, K.; Charbgoo, F.; Abnous, K.; Taghdisi, S.M. Ultrasensitive detection of ochratoxin A using aptasensors. Biosens. Bioelectron. 2017, 98, 168–179. [Google Scholar] [CrossRef]
- Oswald, S.; Karsunke, X.Y.Z.; Dietrich, R.; Märtlbauer, E.; Niessner, R.; Knopp, D. Automated regenerable microarray-based immunoassay for rapid parallel quantification of mycotoxins in cereals. Anal. Bioanal. Chem. 2013, 405, 6405–6415. [Google Scholar] [CrossRef]
- Campone, L.; Piccinelli, A.L.; Celano, R.; Pagano, I.; Russo, M.; Rastrelli, L. Rapid and automated on-line solid phase extraction HPLC–MS/MS with peak focusing for the determination of ochratoxin A in wine samples. Food Chem. 2018, 244, 128–135. [Google Scholar] [CrossRef]
- Meerpoel, C.; Vidal, A.; di Mavungu, J.D.; Huybrechts, B.; Tangni, E.K.; Devreese, M.; Croubels, S.; De Saeger, S. Development and validation of an LC-MS/MS method for the simultaneous determination of citrinin and ochratoxin a in a variety of feed and foodstuffs. J. Chromatogr. 2018, 1580, 100–109. [Google Scholar] [CrossRef] [PubMed]
- Casoni, D.; Badea, M.; Bros, I.; Cobzac, S.C.A. Investigation on image processing parameters for plate evaluation in TLC analysis of mycotoxins. Studia. UBB Chemia. 2017, 62, 89–102. [Google Scholar] [CrossRef]
- Juan, C.; Berrada, H.; Mañes, J.; Oueslati, S. Multi-mycotoxin determination in barley and derived products from Tunisia and estimation of their dietary intake. Food Chem. Toxicol. 2017, 103, 148–156. [Google Scholar] [CrossRef] [PubMed]
- Fujii, S.; Ono, E.Y.S.; Ribeiro, R.M.R.; Assuncao, F.G.A.; Takabayashi, C.R.; de Oliveira, T.; Itano, E.N.; Ueno, Y.; Kawamura, O.; Hirooka, E.Y. A comparison between enzyme immunoassay and HPLC for ochratoxin a detection in green, roasted and instant coffee, Braz. Arch. Biol. Technol. 2007, 50, 349–359. [Google Scholar] [CrossRef]
- Shao, X.; Zhu, L.; Feng, Y.; Zhang, Y.; Luo, Y.; Huang, K.; Xu, W. Detachable nanoladders: A new method for signal identification and their application in the detection of ochratoxin A. Anal. Chim. Acta 2019, 1087, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Zhou, G.; Wang, X.; Zhang, Y.; Li, Z.; Liu, P.; Yu, B.; Zhang, J. A metal-organic framework/aptamer system as a fluorescent biosensor for determination of aflatoxin B1 in food samples. Talanta 2020, 219, 121342. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Hou, Q.; Waterhouse, G.I.N.; Hou, J.; Ai, S.; Li, X. A simple aptamer-based fluorescent aflatoxin B1 sensor using humic acid as quencher. Talanta 2019, 205, 120131. [Google Scholar] [CrossRef]
- Jia, Y.; Wu, F.; Liu, P.; Zhou, G.; Yu, B.; Lou, X.; Xia, F. A label-free fluorescent aptasensor for the detection of Aflatoxin B1 in food samples using AIEgens and graphene oxide. Talanta 2019, 198, 71–77. [Google Scholar] [CrossRef]
- Zhao, Y.; Liu, R.; Sun, W.; Lv, L.; Guo, Z. Ochratoxin A detection platform based on signal amplification by Exonuclease III and fluorescence quenching by gold nano-particles. Sens. Actuator B-Chem. 2018, 255, 1640–1645. [Google Scholar] [CrossRef]
- Jiang, L.; Qian, J.; Yang, X.; Yan, Y.; Liu, Q.; Wang, K.; Wang, K. Amplified impedimetric aptasensor based on gold nanoparticles covalently bound graphene sheet for the picomolar detection of ochratoxin A. Anal. Chim. Acta 2014, 806, 128–135. [Google Scholar] [CrossRef]
- Hu, Z.; Lustig, W.P.; Zhang, J.; Zheng, C.; Wang, H.; Teat, S.J.; Gong, Q.; Rudd, N.D.; Li, J. Effective detection of mycotoxins by a highly luminescent metal−organic framework. J. Am. Chem. Soc. 2015, 137, 16209–16215. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Xia, Y.K.; Chen, M.; Wu, D.Z.; Cai, S.X.; Liu, M.M.; He, W.H.; Chen, J.H. A fluorescent aptasensor based on DNA-scaffolded silver nanoclusters coupling with Zn(II)-ion signal-enhancement for simultaneous detection of OTA and AFB(1). Sens. Actuator B-Chem. 2016, 235, 79–85. [Google Scholar] [CrossRef]
- Wang, B.; Wu, Y.; Chen, Y.; Weng, B.; Xu, L.; Li, C. A highly sensitive aptasensor for OTA detection based on hybridization chain reaction and fluorescent perylene probe. Biosen. Bioelectron. 2016, 81, 125–130. [Google Scholar] [CrossRef] [PubMed]
- Taghdisi, S.M.; Danesh, N.M.; Beheshti, H.R.; Ramezani, M.; Abnous, K. A novel fluorescent aptasensor based on gold and silica nanoparticles for ultrasensitive detection of ochratoxin A. Nanoscale 2016, 8, 2446–3439. [Google Scholar] [CrossRef]
- Hayat, A.; Mishra, R.K.; Catanante, G.; Marty, J.L. Development of an aptasensor based on a fluorescent particles-modified aptamer for ochratoxin A detection. Anal. Bioanal. Chem. 2015, 407, 7815–7822. [Google Scholar] [CrossRef]
- Zhong, Q.; Tian, J.; Wang, B.; Wang, L. PMA based real-time fluorescent LAMP for detection of Vibrio parahaemolyticus in viable but non-culturable state. Food Control 2016, 63, 230–238. [Google Scholar] [CrossRef]
- Xie, S.; Chai, Y.; Yuan, Y.; Bai, L.; Yuan, R. Development of an electrochemical method for Ochratoxin A detection based on aptamer and loop-mediated isothermal amplification. Biosens. Bioelectron. 2014, 55, 324–329. [Google Scholar] [CrossRef]
- Xiong, Q.; Liu, J.; Xie, G.; Mu, D.; Feng, X.; Xu, H. Blocker-tailed PCR coupled with rolling circle amplification for fluorescent detection of emetic Bacillus cereus in milk. LWT-Food Sci. Technol. 2022, 162, 113462. [Google Scholar] [CrossRef]
- Wang, J.; Wang, Y.; Liu, S.; Wang, H.; Zhang, X.; Song, X.; Yu, J.; Huang, J. Primer remodeling amplification-activated multisite-catalytic hairpin assembly enabling concurrent formation of Y-shaped DNA nanotorches for fluorescent assay of ochratoxin A. Analyst 2019, 144, 3389–3397. [Google Scholar] [CrossRef]
- Zeng, R.; Luo, Z.; Su, L.; Zhang, L.; Tang, D.; Niessner, R.; Knopp, D. Palindromic molecular beacon based Z-Scheme BiOCl-Au-CdS photoelectrochemical biodetection. Anal. Chem. 2019, 91, 2447–2454. [Google Scholar] [CrossRef]
- Walker, G.T.; Fraiser, M.S.; Schram, J.L.; Little, M.C.; Nadeau, J.G.; Malinowski, D.P. Strand displacement amplification–an isothermal, in vitro DNA amplification technique. Nucleic Acids Res. 1992, 20, 1691–1696. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Hu, J.; Yang, X.-Y.; Zhang, C. Combination of bidirectional strand displacement amplification with single-molecule detection for multiplexed DNA glycosylases assay. Talanta 2021, 235, 122805. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Xu, X.; Liu, Q.; Wang, L.; Lin, Z.; Chen, G. Ultrasensitive electrochemical biosensor for detection of DNA from Bacillus subtilis by coupling target-induced strand displacement and nicking endonuclease signal amplification. Anal. Chem. 2014, 86, 8785–8790. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Zhang, S.; Li, W.; Zhan, Y.; Yu, L.; Wu, X.; Li, J.; Xu, H.; Yang, G. Label-free and immobilization-free electrochemiluminescent sensing platform for highly sensitive detection of As(III) by combining target-induced strand displacement amplification with polydopamine nanospheres. Sens. Actuat. B-Chem. 2020, 311, 127818. [Google Scholar] [CrossRef]
- Hu, K.; Liu, J.; Chen, J.; Huang, Y.; Zhao, S.; Tian, J.; Zhang, G. An amplified graphene oxide-based fluorescence aptasensor based on target-triggered aptamer hairpin switch and strand-displacement polymerization recycling forbioassays. Biosens. Bioelectron. 2013, 42, 598–602. [Google Scholar] [CrossRef]
- Cruz-Aguado, J.A.; Penner, G. Determination of Ochratoxin A with a DNA Aptamer. J. Agric. Food Chem. 2008, 56, 10456–10461. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Wang, Y.; Marty, J.L.; Yang, X. Aptamer-based colorimetric biosensing of ochratoxin A using unmodified gold nanoparticles indicator. Biosens. Bioelectron. 2011, 26, 2724–2727. [Google Scholar] [CrossRef]
- Yang, Y.-J.; Zhou, Y.; Xing, Y.; Zhang, G.-M.; Zhang, Y.; Zhang, C.-H.; Lei, P.; Dong, C.; Deng, X.; He, Y.; et al. A Label-free aptasensor based on Aptamer/NH2 Janus particles for ultrasensitive electrochemical detection of Ochratoxin A. Talanta 2019, 199, 310–316. [Google Scholar] [CrossRef]
- Li, X.; Falcone, N.; Hossain, M.N.; Kraatz, H.-B.; Chen, X.; Huang, H. Development of a novel label-free impedimetric electrochemical sensor based on hydrogel/chitosan for the detection of ochratoxin A. Talanta 2021, 266, 122183. [Google Scholar] [CrossRef]
- Gökçe, G.; Aissa, S.B.; Nemčeková, K.; Catanante, G.; Raouafi, N.; Marty, J.L. Aptamer-modified pencil graphite electrodes for the impedimetric determination of ochratoxin A. Food Control 2020, 115, 107271. [Google Scholar] [CrossRef]
- Wang, Y.; Fang, Z.; Ning, G.; Mao, S.; Wu, Y.; Wu, S.; Liu, G.-Q. G-quadruplex-bridged triple-helix aptamer probe strategy: A label-free chemiluminescence biosensor for ochratoxin A. Sens. Actuator B-Chem 2019, 298, 126867. [Google Scholar] [CrossRef]





| Oligonucleotide | Sequence (5′→3′) |
|---|---|
| hairpin DNA (HP) | CAGAAGAATCAGGCAATGTTGCCTGTCCGATGCTCCTTTTGATCGGGTGTGGGTGGCGTAAAGGGAGCATCGGACAGGC-spacer |
| Primer | GGAGCATCGGACAG |
| Analysis Method | Linear Range | LOD | Detection Time | Ref |
|---|---|---|---|---|
| Aptamer/NH2 Janus particle-based electrochemical aptasensor | 1 × 10−5–10 nM | 3.3 × 10−3 pM | 30 min | [38] |
| Hydrogel/chitosan-based label-free impedimetric electrochemical sensor | 0.1–100 ng mL−1 | 30 pg mL−1 | 90 min | [39] |
| Fluorescent aptasensor using cascade strand displacement reaction | 1–1000 ng mL−1 | 0.63 ng L−1 | 110 min | [4] |
| Impedimetric aptasensor based on pencil graphite electrodes | 0.1–2.0 ng mL−1 | 0.1 ng L−1 | 90 min | [40] |
| Label-free chemiluminescence biosensor | 0.1–2.0 ng mL−1 | 0.07 ng L−1 | 90 min | [41] |
| Fluorescent aptasensor based on SDA | 0.01–50 ng mL−1 | 0.01 ng mL−1 | 60 min | This study |
| Sample | Added (ng mL−1) | Detected (ng mL−1) | Recovery (%) | RSD (%) | |
|---|---|---|---|---|---|
| Our Method | ELISA | ||||
| wheat | 1.0 | 0.992 | 0.90 | 99.2 | 5.6 |
| 10.0 | 9.6 | 9.52 | 96 | 5.5 | |
| oat | 1.0 | 0.987 | 0.89 | 98.7 | 3.8 |
| 10.0 | 10.01 | 9.89 | 100.1 | 4.9 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, W.; Yang, H.; Zhang, Y.; Wu, H.; Lu, X.; Tan, J.; Zhang, W. A Novel Fluorescent Aptasensor Based on Real-Time Fluorescence and Strand Displacement Amplification for the Detection of Ochratoxin A. Foods 2022, 11, 2443. https://0-doi-org.brum.beds.ac.uk/10.3390/foods11162443
Guo W, Yang H, Zhang Y, Wu H, Lu X, Tan J, Zhang W. A Novel Fluorescent Aptasensor Based on Real-Time Fluorescence and Strand Displacement Amplification for the Detection of Ochratoxin A. Foods. 2022; 11(16):2443. https://0-doi-org.brum.beds.ac.uk/10.3390/foods11162443
Chicago/Turabian StyleGuo, Wei, Haoyu Yang, Yunzhe Zhang, Hao Wu, Xin Lu, Jianxin Tan, and Wei Zhang. 2022. "A Novel Fluorescent Aptasensor Based on Real-Time Fluorescence and Strand Displacement Amplification for the Detection of Ochratoxin A" Foods 11, no. 16: 2443. https://0-doi-org.brum.beds.ac.uk/10.3390/foods11162443