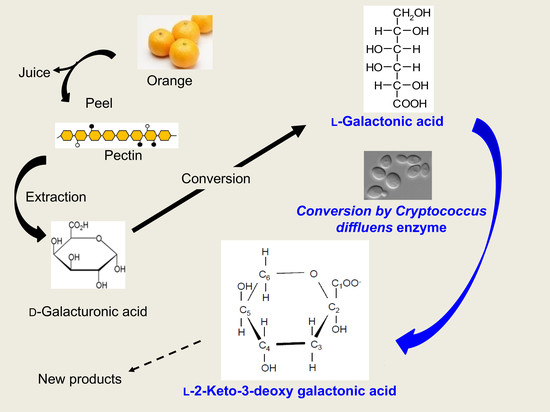
Basidiomycotic Yeast Cryptococcus diffluens Converts l-Galactonic Acid to the Compound on the Similar Metabolic Pathway in Ascomycetes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strain, Media, and Culture Condition
2.2. Preparation of Enzymes
2.2.1. Preparation of Cell-Free Extract
2.2.2. Preparation of Diethylaminoethy (DEAE)-Fractionated Enzyme
2.3. Assay for Sugar Acids by High-Performance Anion-Exchange Chromatography
2.4. Assay for Dehydratase Activity
2.5. Identification of l-2KDGal
2.5.1. Determination of Molecular Mass
2.5.2. Nuclear Magnetic Resonance
3. Results
3.1. Detection of the Product Converted from l-GalA
3.2. Identification of Compound-X
3.2.1. MS Analysis
3.2.2. NMR Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pourbafrani, M.; Forgács, G.; Horváth, I.S.; Niklasson, C.; Taherzadeh, M.J. Production of biofuels, limonene and pectin from citrus wastes. Bioresour. Technol. 2010, 101, 4246–4250. [Google Scholar] [CrossRef] [PubMed]
- Modelska, M.; Berlowska, J.; Kregiel, D.; Cieciura, W.; Antolak, H.; Tomaszewska, J.; Binczarski, M.; Szubiakiewicz, E.; Witonska, I.A. Concept for Recycling Waste Biomass from the Sugar Industry for Chemical and Biotechnological Purposes. Molecules 2017, 22, 1544. [Google Scholar] [CrossRef] [PubMed]
- Doran-Peterson, J.; Cook, M.D.; Brandon, K.S. Microbial conversion of sugars from plant biomass to lactic acid or ethanol. Plant J. 2008, 54, 582–592. [Google Scholar] [CrossRef] [PubMed]
- Grohman, K.; Cameron, R.; Kim, Y.; Widmer, W.; Luzio, G. Extraction and recovery of pectic fragments from citrus processing waste for co-production with ethanol. J. Chem. Technol. Biotechnol. 2013, 88, 395–407. [Google Scholar] [CrossRef]
- Kishida, M.; Seike, Y.; Kawasaki, H. Identification and characterization of a psychrophilic yeast strain newly isolated from the fermentative starter (Loog-pang) of a traditional drink in Thailand. Biocontrol Sci. 2009, 14, 119–122. [Google Scholar] [CrossRef] [PubMed]
- Hamada, S.; Seike, Y.; Tanimori, S.; Sakamoto, T.; Kishida, M. Characterization of d-galacturonate reductase purified from the psychrophilic yeast species Cryptococcus diffluens. J. Biosci. Bioeng. 2011, 111, 518–521. [Google Scholar] [CrossRef] [PubMed]
- Matsubara, T.; Hamada, S.; Wakabayashi, A.; Kishida, M. Production of l-galactonate by recombinant Saccharomyces cerevisiae expressing the galacturonate reductase gene from Cryptococcus diffluens. J. Biosci. Bioeng. 2016, 122, 639–644. [Google Scholar] [CrossRef] [PubMed]
- Hilditch, S.; Berghäll, S.; Kalkkinen, N.; Penttilä, M.; Richard, P. The missing link in the fungal d-galacturonate pathway: Identification of the l-threo-3-deoxy-hexulosonate aldolase. J. Biol. Chem. 2007, 282, 26195–26201. [Google Scholar] [CrossRef] [PubMed]
- Alazi, E.; Khosravi, C.; Homan, T.G.; du Pré, S.; Arentshorst, M.; Falco, M.D.; Pham, T.T.M.; Peng, M.; Aguilar-Pontes, M.V.; Visser, J.; et al. The pathway intermediate 2-keto-3-deoxy-l-galactonate mediates the induction of genes involved in d-galacturonic acid utilization in Aspergillus niger. FEBS Lett. 2017, 591, 1408–1418. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Thiewes, H.; van Kan, J.A.L. The d-galacturonic acid catabolic pathway in Botrytis cinerea. Fungal Genet. Biol. 2011, 48, 990–997. [Google Scholar] [CrossRef] [PubMed]
- Onofri, S.; Barreca, D.; Garuccio, I. Effects of lycorine on growth and effects of l-galactonic acid-γ-lactone on ascorbic acid biosynthesis in strains of Cryptococcus laurentii isolated from Narcissus pseudonarcissus roots and bulbs. Antonie van Leeuwenhoek 2003, 83, 57–61. [Google Scholar] [CrossRef] [PubMed]
- Nishimoto, T.; Furuta, M.; Kataoka, M.; Kishida, M. Important role of catalase in the cellular response of the budding yeast Saccharomyces cerevisiae exposed to ionizing radiation. Curr. Microbiol. 2015, 70, 404–407. [Google Scholar] [CrossRef] [PubMed]
- Buchanan, C.L.; Connaris, H.; Danson, M.J.; Reeve, C.D.; Hough, D.W. An extremely thermostable aldolase from Sulfolobus solfataricus with specificity for non-phosphorylated substrates. Biochem. J. 1999, 343, 563–570. [Google Scholar] [CrossRef] [PubMed]
- Kuorelahti, S.; Jouhten, P.; Maaheimo, H.; Penttilä, M.; Richard, P. l-Galactonate dehydratase is part of the fungal path for d-galacturonic acid catabolism. Mol. Microbiol. 2006, 61, 1060–1068. [Google Scholar] [CrossRef] [PubMed]
- Nishiyama, R.; Inoue, A.; Ojima, T. Identification of 2-keto-3-deoxy-d-gluconate kinase and 2-keto-3-deoxy-d-phosphogluconate aldolase in an alginate-assimilating bacterium, Flavobacterium sp. Strain UMI-01. Mar. Drugs 2017, 15, 37. [Google Scholar] [CrossRef] [PubMed]





| Compound-X A | l-2KDGal B | l-2KDGal C | |
|---|---|---|---|
| Atom | s (ppm) | s (ppm) | s (ppm) |
| C1 | 174.72 | 177.53 | 176.28 |
| C2 | 96.8 | 97.84 | 96.63 |
| C3 | 39.77 | 40.22 | 38.92 |
| C4 | 69.66 | 70.13 | 68.93 |
| C5 | 71.58 | 71.92 | 70.72 |
| C6 | 64.24 | 64.18 | 62.98 |
| H3 | 1.74 | 1.79 | 1.75 |
| H3′ | 2.18 | 2.16 | 2.14 |
| H4 | 3.82 | 3.86 | 3.85 |
| H5 | 3.54 | 3.6 | 3.58 |
| H6 | 3.59 | 3.61 | 3.57 |
| H6′ | 3.77 | 3.8 | 3.78 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Matsubara, T.; Kataoka, M.; Kishida, M. Basidiomycotic Yeast Cryptococcus diffluens Converts l-Galactonic Acid to the Compound on the Similar Metabolic Pathway in Ascomycetes. Fermentation 2019, 5, 73. https://0-doi-org.brum.beds.ac.uk/10.3390/fermentation5030073
Matsubara T, Kataoka M, Kishida M. Basidiomycotic Yeast Cryptococcus diffluens Converts l-Galactonic Acid to the Compound on the Similar Metabolic Pathway in Ascomycetes. Fermentation. 2019; 5(3):73. https://0-doi-org.brum.beds.ac.uk/10.3390/fermentation5030073
Chicago/Turabian StyleMatsubara, Takeo, Michihiko Kataoka, and Masao Kishida. 2019. "Basidiomycotic Yeast Cryptococcus diffluens Converts l-Galactonic Acid to the Compound on the Similar Metabolic Pathway in Ascomycetes" Fermentation 5, no. 3: 73. https://0-doi-org.brum.beds.ac.uk/10.3390/fermentation5030073