Novel Crosstalks between Circadian Clock and Jasmonic Acid Pathway Finely Coordinate the Tradeoff among Plant Growth, Senescence and Defense
Abstract
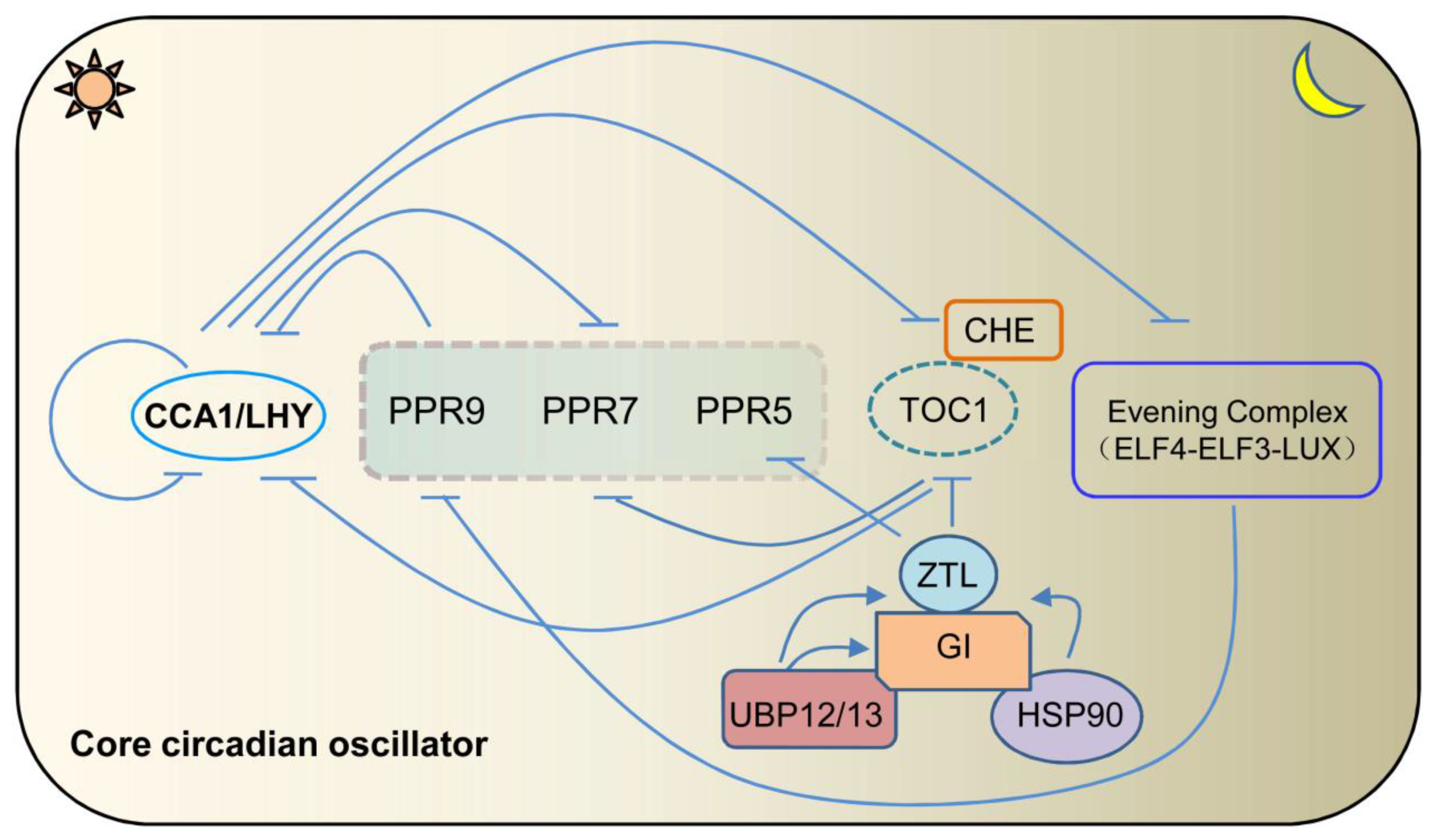
:1. Introduction
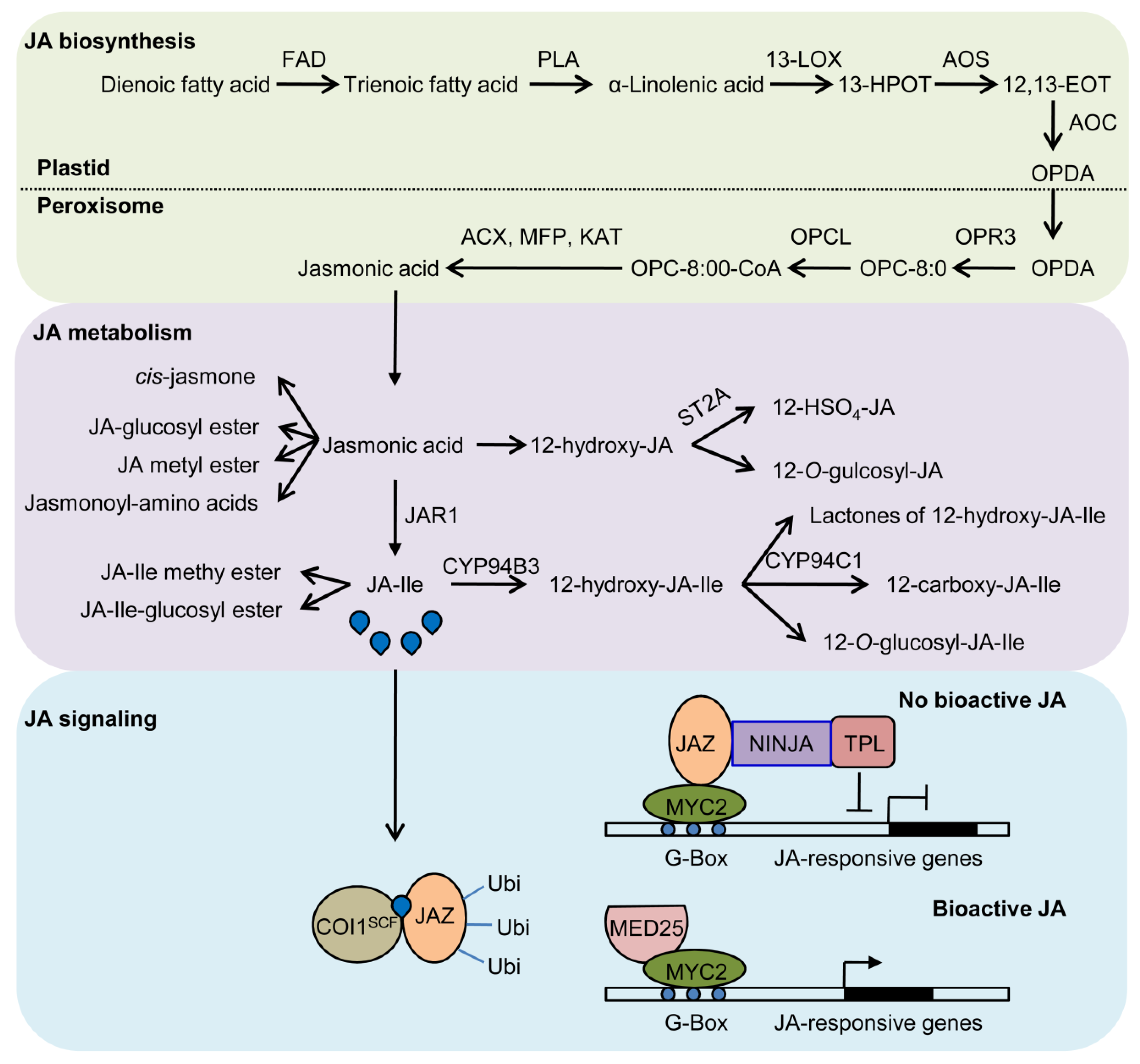
2. Biosynthesis and Metabolism of JA are Regulated by Circadian Clock
3. Circadian Clock Regulates JA-Mediated Plant Development Events
4. Circadian Clock Gates JA Regulated Herbivory Resistance
5. Circadian Clock Gates Temporal Variation Susceptibility to Pathogen by Jasmonates
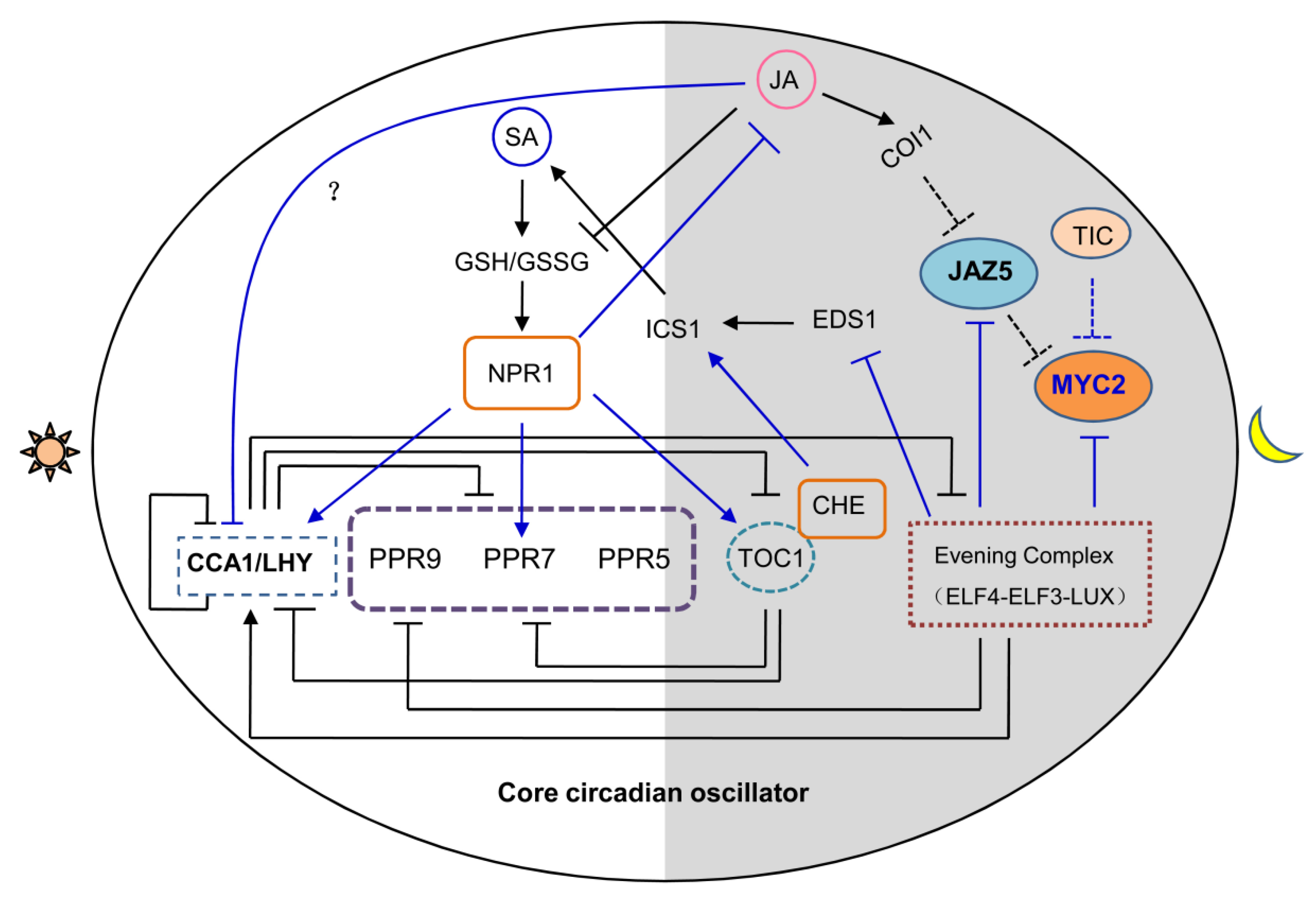
6. Circadian Clock is Associated with the Crosstalk between JA and SA Signaling
7. JA Signaling Reciprocally Affects Clock Activity
8. Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sanchez, S.E.; Kay, S.A. The plant circadian clock: From a simple timekeeper to a complex developmental manager. Cold Spring Harb. Perspect. Biol. 2016, 8. [Google Scholar] [CrossRef]
- Greenham, K.; McClung, C.R. Integrating circadian dynamics with physiological processes in plants. Nat. Rev. Genet. 2015, 16, 598–610. [Google Scholar] [CrossRef]
- Nagel, D.H.; Doherty, C.J.; Pruneda-Paz, J.L.; Schmitz, R.J.; Ecker, J.R.; Kay, S.A. Genome-wide identification of CCA1 targets uncovers an expanded clock network in Arabidopsis. Proc. Natl Acad. Sci. USA 2015, 112, E4802–E4810. [Google Scholar] [CrossRef]
- Harmer, S.L.; Hogenesch, J.B.; Straume, M.; Chang, H.S.; Han, B.; Zhu, T.; Wang, X.; Kreps, J.A.; Kay, S.A. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 2000, 290, 2110–2113. [Google Scholar] [CrossRef]
- Pokhilko, A.; Fernandez, A.P.; Edwards, K.D.; Southern, M.M.; Halliday, K.J.; Millar, A.J. The clock gene circuit in Arabidopsis includes a repressilator with additional feedback loops. Mol. Syst. Biol. 2012, 8. [Google Scholar] [CrossRef]
- Wang, L.; Kim, J.; Somers, D.E. Transcriptional corepressor TOPLESS complexes with pseudoresponse regulator proteins and histone deacetylases to regulate circadian transcription. Proc. Natl. Acad. Sci. USA 2013, 110, 761–766. [Google Scholar] [CrossRef]
- Hicks, K.A.; Albertson, T.M.; Wagner, D.R. EARLY FLOWERING3 encodes a novel protein that regulates circadian clock function and flowering in Arabidopsis. Plant Cell 2001, 13, 1281–1292. [Google Scholar] [CrossRef]
- Doyle, M.R.; Davis, S.J.; Bastow, R.M.; McWatters, H.G.; Kozma-Bognar, L.; Nagy, F.; Millar, A.J.; Amasino, R.M. The ELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana. Nature 2002, 419, 74–77. [Google Scholar] [CrossRef]
- Helfer, A.; Nusinow, D.A.; Chow, B.Y.; Gehrke, A.R.; Bulyk, M.L.; Kay, S.A. LUX ARRHYTHMO encodes a nighttime repressor of circadian gene expression in the Arabidopsis core clock. Curr. Biol. 2011, 21, 126–133. [Google Scholar] [CrossRef]
- Nusinow, D.A.; Helfer, A.; Hamilton, E.E.; King, J.J.; Imaizumi, T.; Schultz, T.F.; Farre, E.M.; Kay, S.A. The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 2011, 475, 398–402. [Google Scholar] [CrossRef]
- Herrero, E.; Kolmos, E.; Bujdoso, N.; Yuan, Y.; Wang, M.; Berns, M.C.; Uhlworm, H.; Coupland, G.; Saini, R.; Jaskolski, M.; et al. EARLY FLOWERING4 recruitment of EARLY FLOWERING3 in the nucleus sustains the Arabidopsis circadian clock. Plant. Cell 2012, 24, 428–443. [Google Scholar] [CrossRef]
- Kim, W.Y.; Fujiwara, S.; Suh, S.S.; Kim, J.; Kim, Y.; Han, L.; David, K.; Putterill, J.; Nam, H.G.; Somers, D.E. ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature 2007, 449, 356–360. [Google Scholar] [CrossRef]
- Baudry, A.; Ito, S.; Song, Y.H.; Strait, A.A.; Kiba, T.; Lu, S.; Henriques, R.; Pruneda-Paz, J.L.; Chua, N.H.; Tobin, E.M.; et al. F-box proteins FKF1 and LKP2 act in concert with ZEITLUPE to control Arabidopsis clock progression. Plant. Cell 2010, 22, 606–622. [Google Scholar] [CrossRef]
- Mas, P.; Kim, W.Y.; Somers, D.E.; Kay, S.A. Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature 2003, 426, 567–570. [Google Scholar] [CrossRef]
- Cha, J.Y.; Kim, J.; Kim, T.S.; Zeng, Q.; Wang, L.; Lee, S.Y.; Kim, W.Y.; Somers, D.E. GIGANTEA is a co-chaperone which facilitates maturation of ZEITLUPE in the Arabidopsis circadian clock. Nat. Commun. 2017, 8. [Google Scholar] [CrossRef]
- Lee, C.M.; Li, M.W.; Feke, A.; Liu, W.; Saffer, A.M.; Gendron, J.M. GIGANTEA recruits the UBP12 and UBP13 deubiquitylases to regulate accumulation of the ZTL photoreceptor complex. Nat. Commun. 2019, 10. [Google Scholar] [CrossRef]
- Nieto, C.; Lopez-Salmeron, V.; Daviere, J.M.; Prat, S. ELF3-PIF4 interaction regulates plant growth independently of the Evening Complex. Curr. Biol. 2015, 25, 187–193. [Google Scholar] [CrossRef]
- Rawat, R.; Schwartz, J.; Jones, M.A.; Sairanen, I.; Cheng, Y.; Andersson, C.R.; Zhao, Y.; Ljung, K.; Harmer, S.L. REVEILLE1, a Myb-like transcription factor, integrates the circadian clock and auxin pathways. Proc. Natl. Acad. Sci. USA 2009, 106, 16883–16888. [Google Scholar] [CrossRef] [Green Version]
- Legnaioli, T.; Cuevas, J.; Mas, P. TOC1 functions as a molecular switch connecting the circadian clock with plant responses to drought. EMBO J. 2009, 28, 3745–3757. [Google Scholar] [CrossRef] [Green Version]
- Goossens, J.; Fernandez-Calvo, P.; Schweizer, F.; Goossens, A. Jasmonates: Signal transduction components and their roles in environmental stress responses. Plant Mol. Biol. 2016, 91, 673–689. [Google Scholar] [CrossRef]
- Zhang, G.; Zhao, F.; Chen, L.; Pan, Y.; Sun, L.; Bao, N.; Zhang, T.; Cui, C.X.; Qiu, Z.; Zhang, Y.; et al. Jasmonate-mediated wound signalling promotes plant regeneration. Nat. Plants 2019, 5, 491–497. [Google Scholar] [CrossRef] [PubMed]
- Kazan, K. Diverse roles of jasmonates and ethylene in abiotic stress tolerance. Trends Plant. Sci. 2015, 20, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Lee, B. Friends or foes: New insights in jasmonate and ethylene co-actions. Plant. Cell Physiol. 2015, 56, 414–420. [Google Scholar] [CrossRef]
- Chini, A.; Gimenez-Ibanez, S.; Goossens, A.; Solano, R. Redundancy and specificity in jasmonate signalling. Curr. Opin. Plant. Biol. 2016, 33, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M.; Laxmi, A. Jasmonates: Emerging players in controlling temperature stress tolerance. Front. Plant. Sci. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Liu, B.; Liu, L.; Song, S. Jasmonate action in plant growth and development. J. Exp. Bot. 2017, 68, 1349–1359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wasternack, C.; Hause, B. Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 2013, 111, 1021–1058. [Google Scholar] [CrossRef]
- Ruan, J.; Zhou, Y.; Zhou, M.; Yan, J.; Khurshid, M.; Weng, W.; Cheng, J.; Zhang, K. Jasmonic acid signaling pathway in plants. Int. J. Mol. Sci. 2019, 20. [Google Scholar] [CrossRef]
- Fonseca, S.; Chini, A.; Hamberg, M.; Adie, B.; Porzel, A.; Kramell, R.; Miersch, O.; Wasternack, C.; Solano, R. (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat. Chem. Biol. 2009, 5, 344–350. [Google Scholar] [CrossRef]
- Fonseca, S.; Chico, J.M.; Solano, R. The jasmonate pathway: The ligand, the receptor and the core signalling module. Curr. Opin. Plant. Biol. 2009, 12, 539–547. [Google Scholar] [CrossRef]
- Xie, D.X.; Feys, B.F.; James, S.; Nieto-Rostro, M.; Turner, J.G. COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 1998, 280, 1091–1094. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Zhang, C.; Gu, M.; Bai, Z.; Zhang, W.; Qi, T.; Cheng, Z.; Peng, W.; Luo, H.; Nan, F.; et al. The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant. Cell 2009, 21, 2220–2236. [Google Scholar] [CrossRef] [PubMed]
- Melotto, M.; Mecey, C.; Niu, Y.; Chung, H.S.; Katsir, L.; Yao, J.; Zeng, W.; Thines, B.; Staswick, P.; Browse, J.; et al. A critical role of two positively charged amino acids in the Jas motif of Arabidopsis JAZ proteins in mediating coronatine- and jasmonoyl isoleucine-dependent interactions with the COI1 F-box protein. Plant. J. 2008, 55, 979–988. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.S.; Niu, Y.; Browse, J.; Howe, G.A. Top hits in contemporary JAZ: An update on jasmonate signaling. Phytochemistry 2009, 70, 1547–1559. [Google Scholar] [CrossRef] [Green Version]
- Pauwels, L.; Goossens, A. The JAZ proteins: A crucial interface in the jasmonate signaling cascade. Plant. Cell 2011, 23, 3089–3100. [Google Scholar] [CrossRef]
- An, C.; Li, L.; Zhai, Q.; You, Y.; Deng, L.; Wu, F.; Chen, R.; Jiang, H.; Wang, H.; Chen, Q.; et al. Mediator subunit MED25 links the jasmonate receptor to transcriptionally active chromatin. Proc. Natl. Acad. Sci. USA 2017, 114, E8930–E8939. [Google Scholar] [CrossRef] [Green Version]
- Goossens, J.; Mertens, J.; Goossens, A. Role and functioning of bHLH transcription factors in jasmonate signalling. J. Exp. Bot. 2017, 68, 1333–1347. [Google Scholar] [CrossRef]
- Qi, T.; Wang, J.; Huang, H.; Liu, B.; Gao, H.; Liu, Y.; Song, S.; Xie, D. Regulation of jasmonate-induced leaf senescence by antagonism between bHLH subgroup IIIe and IIId factors in Arabidopsis. Plant. Cell 2015, 27, 1634–1649. [Google Scholar] [CrossRef]
- Zhu, X.; Chen, J.; Xie, Z.; Gao, J.; Ren, G.; Gao, S.; Zhou, X.; Kuai, B. Jasmonic acid promotes degreening via MYC2/3/4- and ANAC019/055/072-mediated regulation of major chlorophyll catabolic genes. Plant. J. 2015, 84, 597–610. [Google Scholar] [CrossRef]
- Wang, H.; Li, Y.; Pan, J.; Lou, D.; Hu, Y.; Yu, D. The bHLH transcription factors MYC2, MYC3, and MYC4 are required for jasmonate-mediated inhibition of flowering in Arabidopsis. Mol. Plant. 2017, 10, 1461–1464. [Google Scholar] [CrossRef]
- Wang, W.; Barnaby, J.Y.; Tada, Y.; Li, H.; Tor, M.; Caldelari, D.; Lee, D.U.; Fu, X.D.; Dong, X. Timing of plant immune responses by a central circadian regulator. Nature 2011, 470, 110–114. [Google Scholar] [CrossRef] [PubMed]
- Covington, M.F.; Maloof, J.N.; Straume, M.; Kay, S.A.; Harmer, S.L. Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol. 2008, 9. [Google Scholar] [CrossRef]
- Michael, T.P.; Mockler, T.C.; Breton, G.; McEntee, C.; Byer, A.; Trout, J.D.; Hazen, S.P.; Shen, R.; Priest, H.D.; Sullivan, C.M.; et al. Network discovery pipeline elucidates conserved time-of-day-specific cis-regulatory modules. PLoS Genet. 2008, 4. [Google Scholar] [CrossRef] [PubMed]
- Sauerbrunn, N.; Schlaich, N.L. PCC1: A merging point for pathogen defence and circadian signalling in Arabidopsis. Planta 2004, 218, 552–561. [Google Scholar] [CrossRef] [PubMed]
- Goodspeed, D.; Chehab, E.W.; Min-Venditti, A.; Braam, J.; Covington, M.F. Arabidopsis synchronizes jasmonate-mediated defense with insect circadian behavior. Proc. Natl. Acad. Sci. USA 2012, 109, 4674–4677. [Google Scholar] [CrossRef] [PubMed]
- Yamashino, T.; Kitayama, M.; Mizuno, T. Transcription of ST2A encoding a sulfotransferase family protein that is involved in jasmonic acid metabolism is controlled according to the circadian clock- and PIF4/PIF5-mediated external coincidence mechanism in Arabidopsis thaliana. Biosci. Biotech. Bioch. 2013, 77, 2454–2460. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, Y.; Wei, H.; Li, N.; Tian, W.; Chong, K.; Wang, L. Circadian evening complex represses jasmonate-induced leaf senescence in Arabidopsis. Mol. Plant. 2018, 11, 326–337. [Google Scholar] [CrossRef]
- Nitschke, S.; Cortleven, A.; Iven, T.; Feussner, I.; Havaux, M.; Riefler, M.; Schmulling, T. Circadian stress regimes affect the circadian clock and cause jasmonic acid-dependent cell death in cytokinin-deficient Arabidopsis plants. Plant. Cell 2016, 28, 1616–1639. [Google Scholar] [CrossRef]
- Shin, J.; Heidrich, K.; Sanchez-Villarreal, A.; Parker, J.E.; Davis, S.J. TIME FOR COFFEE represses accumulation of the MYC2 transcription factor to provide time-of-day regulation of jasmonate signaling in Arabidopsis. Plant Cell 2012, 24, 2470–2482. [Google Scholar] [CrossRef]
- Ameye, M.; Allmann, S.; Verwaeren, J.; Smagghe, G.; Haesaert, G.; Schuurink, R.C.; Audenaert, K. Green leaf volatile production by plants: A meta-analysis. New Phytol 2018, 220, 666–683. [Google Scholar] [CrossRef]
- Joo, Y.; Schuman, M.C.; Goldberg, J.K.; Wissgott, A.; Kim, S.G.; Baldwin, I.T. Herbivory elicits changes in green leaf volatile production via jasmonate signaling and the circadian clock. Plant. Cell Environ. 2019, 42, 972–982. [Google Scholar] [CrossRef] [PubMed]
- Yon, F.; Joo, Y.; Cortes Llorca, L.; Rothe, E.; Baldwin, I.T.; Kim, S.G. Silencing Nicotiana attenuata LHY and ZTL alters circadian rhythms in flowers. New Phytol. 2016, 209, 1058–1066. [Google Scholar] [CrossRef]
- Mithofer, A.; Boland, W. Plant defense against herbivores: Chemical aspects. Annu. Rev. Plant. Biol. 2012, 63, 431–450. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Llorca, L.C.; Schuman, M.C.; Wang, Y.; Wang, L.; Joo, Y.; Wang, M.; Vassao, D.G.; Baldwin, I.T. ZEITLUPE in the roots of wild tobacco regulates jasmonate-mediated nicotine biosynthesis and resistance to a generalist herbivore. Plant. Physiol. 2018, 177, 833–846. [Google Scholar] [CrossRef]
- Bhardwaj, V.; Meier, S.; Petersen, L.N.; Ingle, R.A.; Roden, L.C. Defence responses of Arabidopsis thaliana to infection by Pseudomonas syringae are regulated by the circadian clock. PLoS ONE 2011, 6. [Google Scholar] [CrossRef] [PubMed]
- Korneli, C.; Danisman, S.; Staiger, D. Differential control of pre-invasive and post-invasive antibacterial defense by the Arabidopsis circadian clock. Plant. Cell Physiol. 2014, 55, 1613–1622. [Google Scholar] [CrossRef]
- Zhang, C.; Gao, M.; Seitz, N.C.; Angel, W.; Hallworth, A.; Wiratan, L.; Darwish, O.; Alkharouf, N.; Dawit, T.; Lin, D.; et al. LUX ARRHYTHMO mediates crosstalk between the circadian clock and defense in Arabidopsis. Nat. Commun. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Karapetyan, S.; Dong, X. Redox and the circadian clock in plant immunity: A balancing act. Free Radical Biol. Med. 2018, 119, 56–61. [Google Scholar] [CrossRef]
- Wang, G.; Zhang, C.; Battle, S.; Lu, H. The phosphate transporter PHT4;1 is a salicylic acid regulator likely controlled by the circadian clock protein CCA1. Front. Plant. Sci. 2014, 5. [Google Scholar] [CrossRef] [Green Version]
- Zheng, X.Y.; Zhou, M.; Yoo, H.; Pruneda-Paz, J.L.; Spivey, N.W.; Kay, S.A.; Dong, X. Spatial and temporal regulation of biosynthesis of the plant immune signal salicylic acid. Proc. Natl. Acad. Sci. USA 2015, 112, 9166–9173. [Google Scholar] [CrossRef] [Green Version]
- Tada, Y.; Spoel, S.H.; Pajerowska-Mukhtar, K.; Mou, Z.; Song, J.; Wang, C.; Zuo, J.; Dong, X. Plant immunity requires conformational changes of NPR1 via S-nitrosylation and thioredoxins. Science 2008, 321, 952–956. [Google Scholar] [CrossRef] [PubMed]
- Mou, Z.; Fan, W.; Dong, X. Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 2003, 113, 935–944. [Google Scholar] [CrossRef]
- Santner, A.; Calderon-Villalobos, L.I.; Estelle, M. Plant hormones are versatile chemical regulators of plant growth. Nat. Chem. Biol. 2009, 5, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Roldan, V.; Fermas, S.; Brewer, P.B.; Puech-Pages, V.; Dun, E.A.; Pillot, J.P.; Letisse, F.; Matusova, R.; Danoun, S.; Portais, J.C.; et al. Strigolactone inhibition of shoot branching. Nature 2008, 455, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Hagihara, S.; Yamada, R.; Itami, K.; Torii, K.U. Dissecting plant hormone signaling with synthetic molecules: Perspective from the chemists. Curr. Opin. Plant. Biol. 2019, 47, 32–37. [Google Scholar] [CrossRef] [PubMed]
- Larrieu, A.; Vernoux, T. Comparison of plant hormone signalling systems. Essays Biochem. 2015, 58, 165–181. [Google Scholar] [CrossRef] [PubMed]
- Hanano, S.; Domagalska, M.A.; Nagy, F.; Davis, S.J. Multiple phytohormones influence distinct parameters of the plant circadian clock. Genes Cells 2006, 11, 1381–1392. [Google Scholar] [CrossRef]
- Zhou, M.; Wang, W.; Karapetyan, S.; Mwimba, M.; Marques, J.; Buchler, N.E.; Dong, X. Redox rhythm reinforces the circadian clock to gate immune response. Nature 2015, 523, 472–476. [Google Scholar] [CrossRef]
- Kim, H.; Kim, H.J.; Vu, Q.T.; Jung, S.; McClung, C.R.; Hong, S.; Nam, H.G. Circadian control of ORE1 by PRR9 positively regulates leaf senescence in Arabidopsis. Proc. Natl. Acad. Sci. USA 2018, 115, 8448–8453. [Google Scholar] [CrossRef]
- Mao, Y.B.; Liu, Y.Q.; Chen, D.Y.; Chen, F.Y.; Fang, X.; Hong, G.J.; Wang, L.J.; Wang, J.W.; Chen, X.Y. Jasmonate response decay and defense metabolite accumulation contributes to age-regulated dynamics of plant insect resistance. Nat. Commun. 2017, 8. [Google Scholar] [CrossRef]
- Grundy, J.; Stoker, C.; Carre, I.A. Circadian regulation of abiotic stress tolerance in plants. Front. Plant. Sci. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Bo, C.; Wang, L. Novel Crosstalks between Circadian Clock and Jasmonic Acid Pathway Finely Coordinate the Tradeoff among Plant Growth, Senescence and Defense. Int. J. Mol. Sci. 2019, 20, 5254. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20215254
Zhang Y, Bo C, Wang L. Novel Crosstalks between Circadian Clock and Jasmonic Acid Pathway Finely Coordinate the Tradeoff among Plant Growth, Senescence and Defense. International Journal of Molecular Sciences. 2019; 20(21):5254. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20215254
Chicago/Turabian StyleZhang, Yuanyuan, Cunpei Bo, and Lei Wang. 2019. "Novel Crosstalks between Circadian Clock and Jasmonic Acid Pathway Finely Coordinate the Tradeoff among Plant Growth, Senescence and Defense" International Journal of Molecular Sciences 20, no. 21: 5254. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20215254




