1. Introduction
Steroid hormone receptors are ligand-dependent transcription factors that require the dynamic and highly ordered assembly of multiple chaperone and cochaperone heterocomplexes to reach a functional conformation [
1]. At least twelve proteins in at least three distinct complexes contribute to receptor folding and/or holding within the cytoplasm. The mature complex in which the receptor is capable of high-affinity hormone-binding includes the 90-kDa heat shock protein (Hsp90), the Hsp90-associated 23-kDA cochaperone (p23), and one of a family of tetratricopeptide repeat (TPR)-containing proteins termed immunophilins. Unlike most of the Hsp90-associated cochaperones, the TPR-containing cochaperones that present in the mature receptor-Hsp90 complex are functionally specific for a subset of Hsp90 client proteins [
2]. As a result, Hsp90 is thought to sample the available pool of TPR-containing cochaperones, and the client protein present in the complex determines which TPR-containing cochaperone will functionally interact.
The 52-kDa FK506-binding protein (FKBP52) associates with receptor-Hsp90 complexes by way of a C-terminal TPR domain [
3,
4] and is a specific positive regulator of androgen receptor (AR), glucocorticoid receptor (GR), and progesterone receptor (PR) signaling in cellular assays [
5,
6,
7]. These results have been confirmed in animals as
fkbp52-deficient (52KO) mice display phenotypes related to reduced AR, PR, and GR signaling [
6,
8]. The specific mechanism by which FKBP52 functionally influences receptor activity is unknown, but it is likely that FKBP52 participates in multiple distinct steps within the receptor signaling pathways. Data suggest that FKBP52 not only influences receptor hormone-binding affinity within the mature Hsp90 complex but also participates in hormone-induced receptor translocation to the nucleus through direct interactions with the dynein motor protein [
9,
10]. In addition, FKBP52 was recently demonstrated to interact directly with β-catenin to promote β-catenin interaction with, and upregulation of AR activity [
11]. Interestingly, this mechanism was independent of FKBP52 binding to Hsp90. Despite the multiple roles identified for FKBP52, it is clear that FKBP52 is a functionally critical factor for AR, GR, and PR activity at physiological hormone concentrations in vivo.
Studies with chimeric receptor proteins demonstrated that FKBP52-mediated effects localized to the receptor hormone-binding domain [
5]. In support of this idea, we previously demonstrated that mutations on the AR binding function 3 (BF3) surface cause increased dependence (also termed FKBP52 hypersensitivity) on FKBP52 for function [
6,
12]. We also identified and characterized a series of small molecules that specifically inhibit FKBP52-mediated potentiation of AR signaling, and available evidence suggests that these molecules target the AR BF3 surface [
12]. Taken together, these findings suggest that the AR BF3 surface is an FKBP52 interaction and/or regulatory surface. The Bag-1L protein was also recently found to regulate AR activity through the BF3 surface, suggesting that the AR BF3 surface may serve as a promiscuous regulatory surface for several factors, including FKBP52 [
13]. Mutations within the H1–H3 loop in the GR hormone-binding domain were also shown to influence FKBP52-mediated receptor activity [
14]. The H1–H3 loop may act as a regulatory surface that promotes allosteric conformational changes in BF3, given that FKBP52 does not directly interact at the H1–H3 loop. However, evidence suggests that the co-regulation of AR by FKBP52 and β-catenin through the AR BF3 surface is a specific AR regulatory mechanism [
11,
15]. Thus, FKBP52 may influence the activity of multiple receptors through multiple distinct sites, including the H1–H3 loop.
A detailed understanding of the residues and domains that contribute to the FKBP52 function can inform ongoing efforts to therapeutically target FKBP52 for the disruption of receptor activity. FKBP52 shares approximately 70% sequence similarity to FKBP51, yet FKBP51 overexpression does not potentiate receptor function in our yeast and mammalian cell assay systems. Thus, this functional difference was exploited in previous studies to map the domains and residues on FKBP52 that are required for function [
5,
16]. The FKBP52 N-terminal FK506 binding domain (FK1) is required for receptor regulation, and functional domain mapping studies demonstrated that two mutations (A116V and L119P), combined were sufficient to confer full receptor potentiating activity to FKBP51. The analogous residues on FKBP52 are within a proline-rich loop (also termed 80 s loop or β4–β5 loop) overhanging the peptidyl-prolyl
cis/
trans isomerase (PPIase) catalytic pocket, suggesting that the proline-rich loop is critical for function. PPIase enzymatic activity is not required for function, but the FKBP ligand and PPIase inhibitor FK506 disrupts receptor potentiation, possibly through the disruption of FK1 and/or proline-rich loop interactions within receptor complexes. Although this region was identified as important for FKBP52 function, the converse mutations in FKBP52 only reduced potentiation by approximately 40%. Thus, the FK1 proline-rich loop is necessary for normal FKBP52 function, but it is not sufficient alone to generate a full response. Thus, other residues likely exist within the FKBP52 FK1 and/or FK2 domains that are required for full FKBP52 function. This is further supported by the lack of receptor potentiating activity of an FKBP52 ortholog from zebra fish.
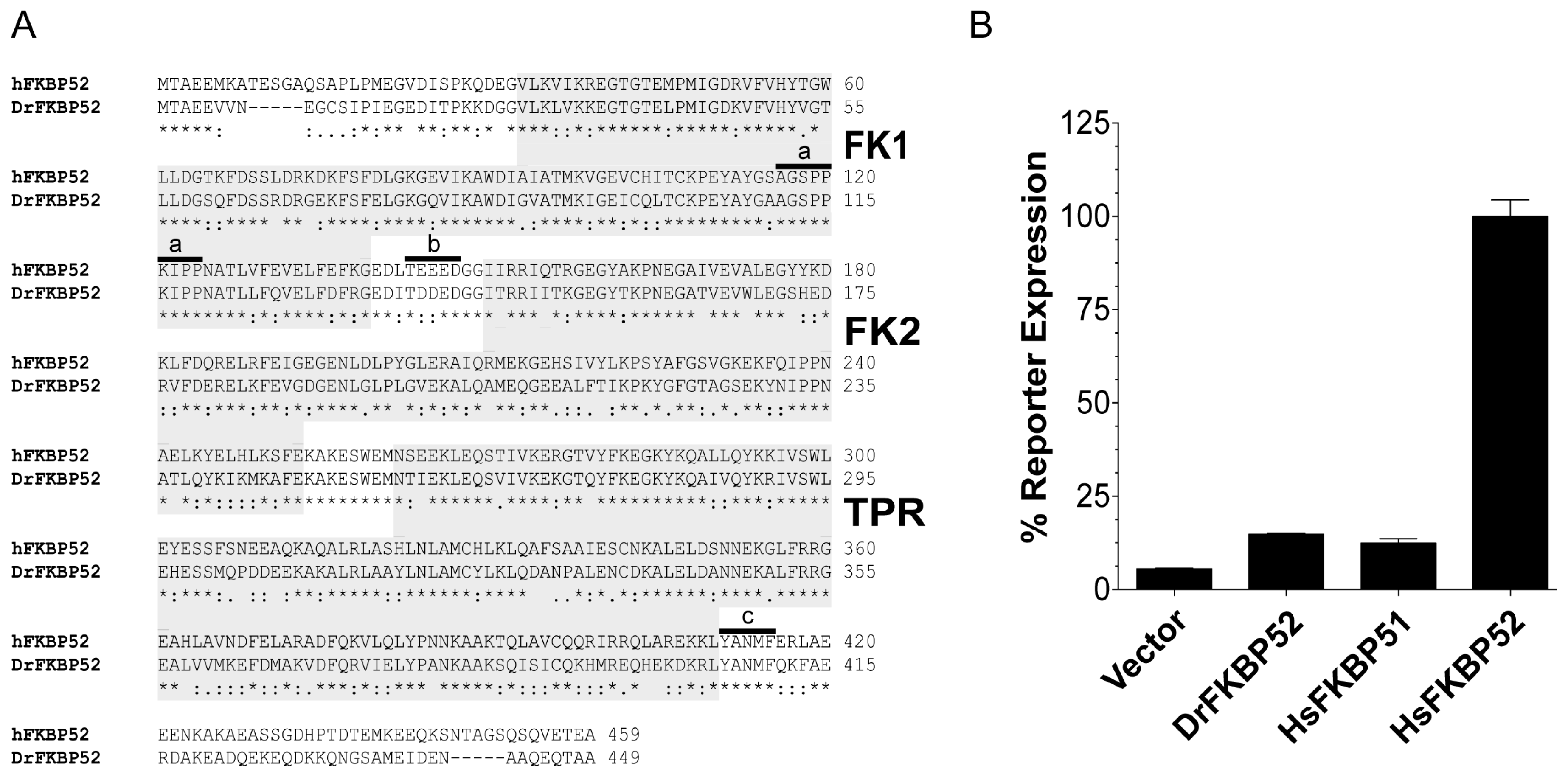
Zebra fish (Danio rerio; Dr) FKBP52 is identical to human (Homo sapiens; Hs) FKBP52 with respect to all known residues that are critical for receptor potentiation. However, as detailed here, DrFKBP52 fails to potentiate receptor activity in reporter assays. Taking advantage of this functional difference, the genetic selection of gain-of-function random DrFKBP52 mutants was used to identify and characterize additional residues and/or domains that are critical for FKBP52 potentiation of receptor activity.
3. Discussion
Human FKBP52 is an Hsp90 cochaperone that positively regulates hormone-dependent transcriptional activity of AR, GR, and PR. Its physiological relevance has been established in studies showing impaired reproductive development of male and female
fkbp52-deficient mice [
6,
7,
8,
20,
21]. While the specific mechanisms for receptor potentiation are still poorly understood, studies implicate FKBP52 in the regulation of receptor–ligand binding and trafficking (reviewed in [
10]). FKBP52 assembles with receptor complexes through its association with Hsp90 to exert effects on receptor activity. In particular, the N-terminal FK1 PPIase domain of FKBP52 has been recognized as important for activity [
16]. While the PPIase enzymatic activity was not required for FKBP52 regulation of receptor activity, these studies highlighted the importance of the proline-rich loop within the FK1 domain. Introducing both the A116V and L119P mutations in the FKBP51 proline-rich loop conferred full receptor potentiating ability similar to that seen with wild type FKBP52. While the L119P mutation made FKBP51 more like FKBP52 at this site, the significance of the A116V mutation was still unclear given that FKBP52 also has an alanine at this position. In addition to the proline-rich loop, studies suggested that the short FK-linker between the FK1 and FK2 domains behaves as a flexible hinge region that may facilitate non-covalent interactions between the FK1 and FK2 domains, revealing a potential role for the FK2 domain in facilitating a functional FK1 conformation [
5,
22,
23].
To further elucidate the domain and residue requirements for FKBP52 potentiation of receptor activity, we took a cross-species comparative approach. Despite the presence of the proline-rich loop, FK linker, Hsp90 binding motif, and predicted structural similarities to HsFKBP52, DrFKBP52 still does not potentiate receptor activity (
Figure 1). Taking advantage of this functional distinction, DrFKBP52 was randomly mutated, and a yeast selection screen was designed to isolate DrFKBP52 mutants that gain receptor potentiation activity (
Figure 2). The gain-of-function mutants that were repeatedly identified were A111V (nine times) and T157R (seven times) (
Figure 3). Separately these mutants had sufficient activity to increase potentiation of AR activity three- to five-fold as compared to wild type DrFKBP52 (
Figure 4). Interestingly, the DrFKBP52-A111V/T157R double mutant completely restored activity to levels similar to HsFKBP52 in both yeast and mammalian cells. It should be noted that while the T157A mutant was able to confer receptor potentiation activity separately, combining this mutation with A111V did not restore full activity as compared to HsFKBP52. Given the flexibility of the hinge region, these data suggest a possible salt bridge between the FK1 and FK2 domains may specifically affect FKBP52 regulation of steroid hormone receptor activity. Interestingly, T157 is located down-stream of IRRIQTR, a highly positively charged region that, like the proline-rich loop region, is highly conserved in FKBP52 and FKBP51 across species, though some are more positively charged in this region. Though we have not confirmed a specific residue in the FK2 domain that is important for HsFKBP52 potentiation, we postulate that residues near the FK-linker region may facilitate FK1-FK2 interactions that allosterically affect FK1 proline-rich loop conformation, as previous studies suggest [
18]. The T157R mutation in Dr-FKBP52 could be enhancing such an interaction between FK1 and FK2.
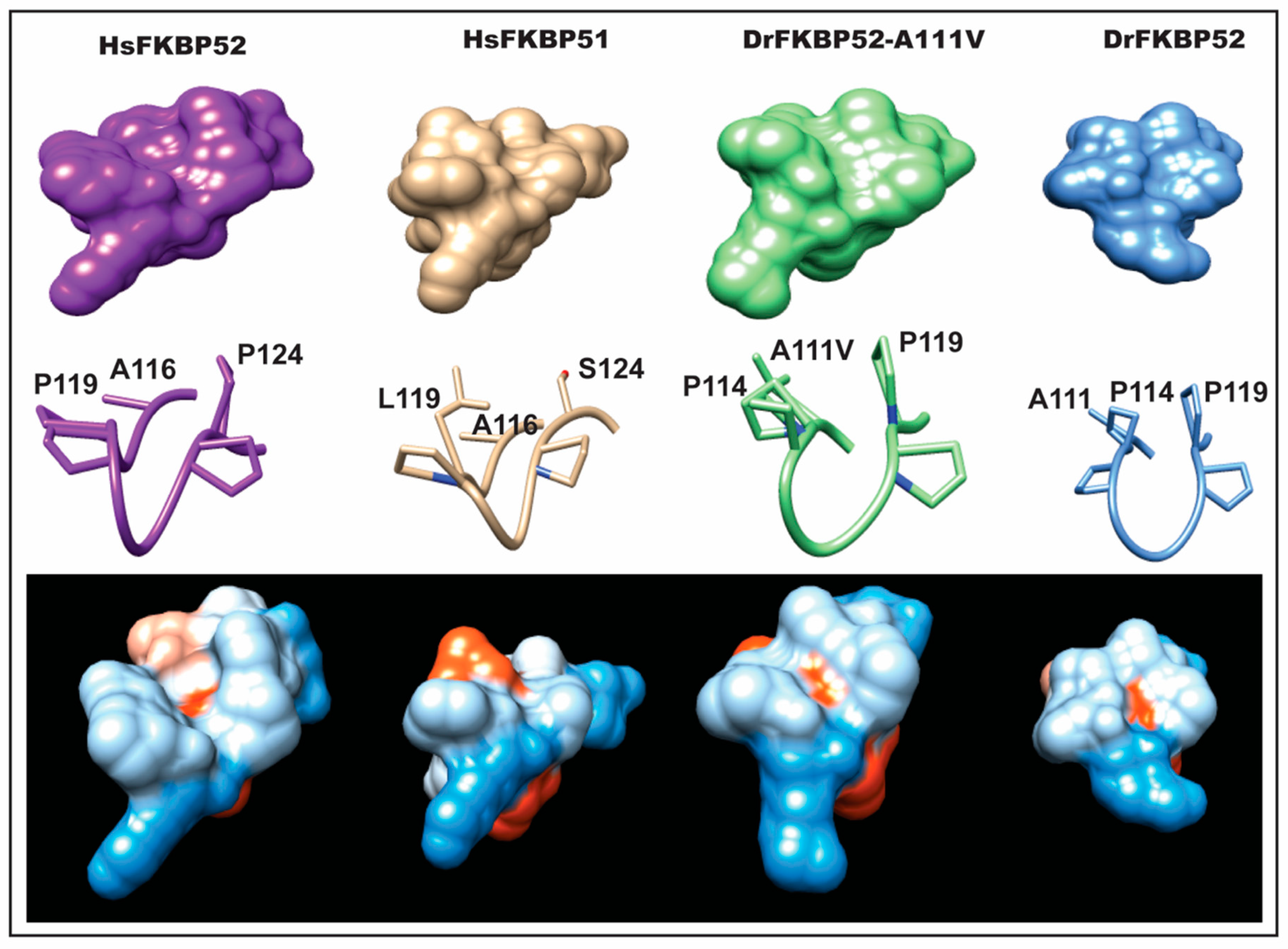
Interestingly, A111 in DrFKBP52 is analogous to A116 in HsFKBP51 and HsFKBP52, a proline-rich loop residue previously highlighted by Riggs et al. [
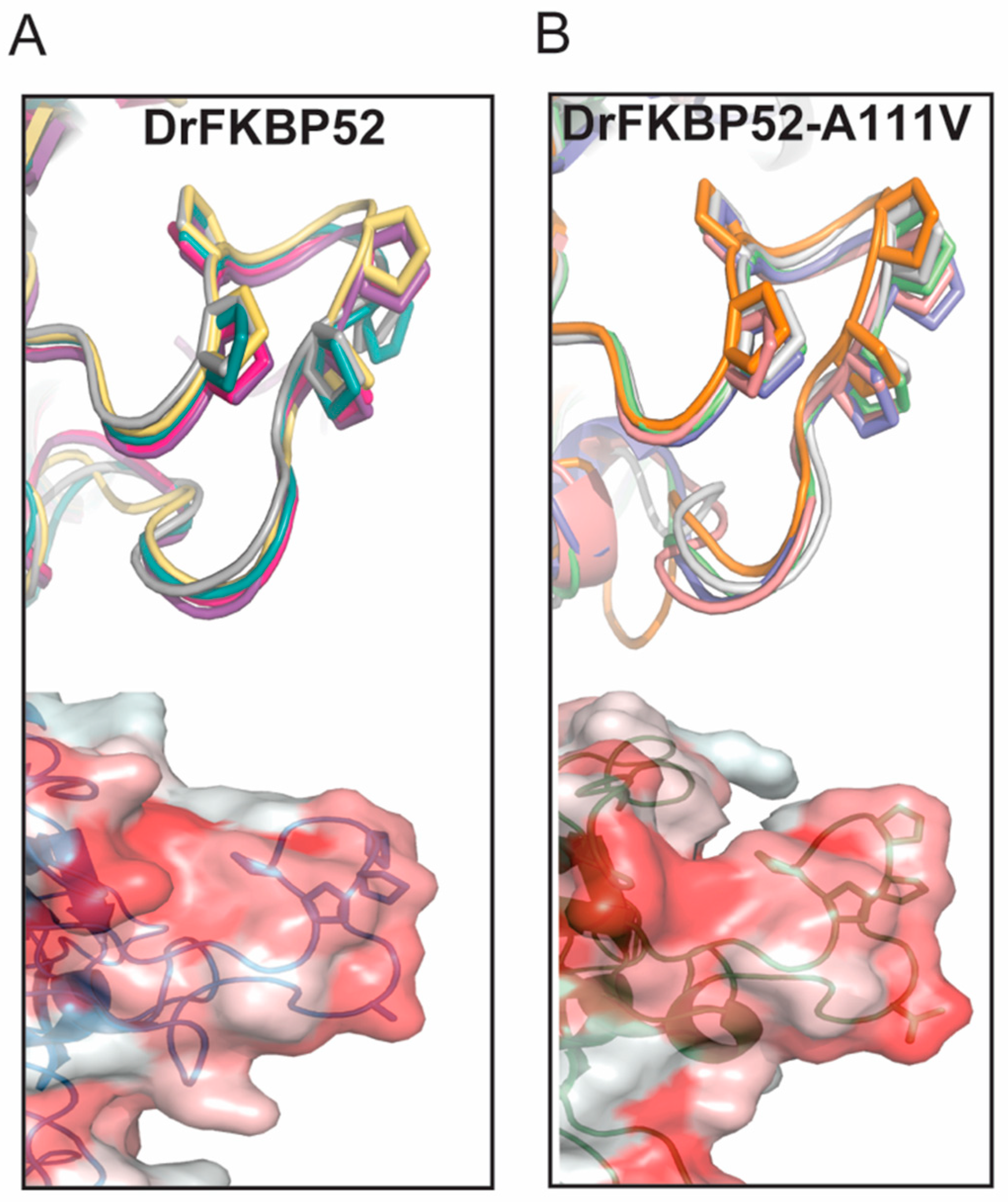
16]. Because wild type DrFKBP52, HsFKBP51, and HsFKBP52 all contain alanine at loop position 111/116, we performed homology modeling to predict the role of this amino acid position may play in determining the proline-rich loop structure (
Figure 5 and
Figure 6). A comparison of the corresponding loops in HsFKBP52, HsFKBP51, DrFKBP52, and DrFKBP52-A111V revealed a predicted structural difference due to the respective amino acids at position 111/116. The DrFKBP52 proline side chains (P114 and P119) protrude into a hydrophobic notch that forms along the top of the loop, more similar to HsFKBP51. The projection of these two residues is significantly altered by the addition of valine at position 111 in DrFKBP52. The conformation of the analogous prolines in DrFKBP52-A111V spread more outward, similar to HsFKBP52. Hydrophobic surface depictions show that HsFKBP52, HsFKBP51, DrFKBP52, and DrFKBP52-A111V all possess a hydrophobic notch on the surface of the proline loop. This surface is blocked by the projection of surrounding residues in DrFKBP52 and HsFKBP51, both of which lack receptor potentiation activity. Only HsFKBP52 and DrFKBP52-A111V, both of which potentiate receptor activity, retain an open docking conformation compatible for protein–protein interaction. These predictive data suggest that DrFKBP52-A111V, in a manner comparable to HsFKBP52, forms a functionally important contact surface via the FK1 hydrophobic notch with other components within the heterocomplex. Human FKBP51 and DrFKBP52 lack this contact due to the altered loop conformation imposed by occluding leucine and proline, inhibiting adequate protein–protein interaction.
We propose that the HsFKBP52 proline-rich loop, also referred to as the β4–β5 loop, via the open conformation that exposes the hydrophobic notch, forms a specific contact with the receptor ligand-binding domain (LBD). This contact augments the steroid receptor response to hormone. We know that HsFKBP52 binds Hsp90 directly, and through this interaction, selectively potentiates the receptors. Additionally, studies have shown that FKBP52-dependent potentiation is localized in the receptor LBD [
5,
23]. Since HsFKBP52 selectively potentiates the hormone response in AR, GR, and PR, but not the activity of the estrogen receptor, we conclude that the relevant FK1 interaction for potentiation is receptor-specific, and not exclusively through Hsp90. We presume that the hydrophobic notch that forms above the proline loop region could allow for a more efficient interaction, possibly with the receptor LBD. Through this study, we conclude that the proline residues in the loop are not the sole indicator of functionality, but the overall conformation of the proline-rich loop is critical to maintaining FKBP52 potentiation of the steroid hormone receptor activity.
4. Materials and Methods
Yeast strains and β-galactosidase reporter assays. For AR-dependent reporter assays [
5], the strains were co-transformed with the indicated plasmids including an AR-dependent β-galactosidase reporter plasmid (pUCΔss-26X), and a high-copy number glyceraldehyde phosphate dehydrogenase (GPD) promoter-regulated yeast expression vector carrying the indicated steroid hormone receptor, HsFKBP51, HsFKBP52, DrFKBP52, and/or site-directed mutants where indicated. Human AR and the mutant AR-P723S were cloned, respectively, into p424GPD and p424TEF [
24]. Dihydrotestosterone (DHT) was obtained from Sigma (St. Louis, MO, USA), and dilutions in ethanol were set up to avoid ethanol vehicle concentrations above 1% in the yeast cultures. Hormone-induced reporter activity was measured from yeast extracts as described previously with a single two-hour time point measurement. The data were normalized to cell density by dividing relative light units (RLU) by the optical density (OD) of the cultures at 600 nm (RLU/OD600) and further normalized to percentages. All data shown were generated from at least three biological and technical replicates.
Selection for gain-of-function DrFKBP52 mutants. Error-prone PCR (GeneMorphII; Stratagene, La Jolla, CA, USA) with conditions recommended to produce low-frequency mutagenesis (50–100ng template DNA per reaction) was used to generate the DrFKBP52 mutant libraries using p425GPD-DrFKBP52 as a template. The selection strain YNK435 (MATa: ura3-52, lys2-801, ade2-101, trp1-63, his3-200, leu2-1, pdr5::GT3Z, his3::GT3H, p425TEF-AR-P723S) was then co-transformed with 400 ng of the PCR product and 100 ng of linearized p424GPD expression vector. Gain-of-function mutants were identified by colony growth on synthetic complete medium lacking tryptophan, leucine, and histidine (SC-WLH) supplemented with 10 nM DHT and 10 mM 3-amino-1,2,4-triazole (3-AT). The concentrations of both DHT and 3-AT were chosen based on concentrations that produced the maximum difference in growth between strains expressing either HsFKBP52 or DrFKBP52.The colonies containing DrFKBP52 that grew comparable to those containing HsFKBP52 were selected. The gain-of-function phenotype was then confirmed by assessing the ability to potentiate AR-dependent reporter gene expression in the selection strain. The DrFKBP52 mutant plasmids were then extracted from yeast lysates using standard procedures, and the extracted mutant plasmids were transformed into strain W303a expressing wild type AR and an AR-responsive β-galactosidase reporter plasmid (pUCΔss-26X). Transformants were assayed for potentiation of hormone signaling, as described above. Those mutated DrFKBP52 genes that retained the ability to potentiate AR activity in this clean genetic background were then sequenced. The identified mutations were then put in individually and in combination into the DrFKBP52 gene by site-directed mutagenesis to determine the mutation or combination of mutations that confer the gain-of-function. Mutations were introduced by site-directed mutagenesis (QuikChange II XL; Stratagene, La Jolla, CA, USA) into the HsFKBP52 or DrFKBP52 genes directly in the yeast expression vector (p423GPD).
Receptor-mediated reporter assays in mouse embryonic fibroblasts. Mouse embryonic fibroblasts derived from FKBP52 knockout mice (52KO MEF) [
6] were cultured in 5% CO2 in HyClone minimal essential media/Eagles essential salt solution with 2 mM L-glutamine (Thermal Scientific, Logan, UT, USA) supplemented with 10% charcoal/dextran treated fetal bovine serum (FBS) (HyClone, Logan, UT, USA) 24 h prior to the transfection. Cells were cultured in 6-well plates until they were 80% confluent. They were transfected in triplicate using lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. Transfections were performed for 5 h at a DNA to lipofectamine ratio of 1:3 in MEM-EBSS lacking FBS. The transfection cocktail was mixed as follows: 50 ng of the constitutive β-galactosidase reporter plasmid (pCMVβ; Clonetech, Mountain View, CA, USA), 400 ng of the firefly luciferase reporter driven by the androgen-dependent Probasin promoter (pT81; American Type Culture Collection, Manassas, VA, USA), and 800 ng of the pCI-Neo plasmid (Promega, Madison, WI, USA) with or without expression of human AR, HsFKBP52, DrFKBP52, and/or DrFKBP52 mutants. Twenty-four hours after transfection, the medium was replaced with medium containing 10 pM DHT, or a range of doses as indicated. Sixteen hours after hormone addition, cells in each well were lysed using 100 μL mammalian protein extraction reagent (M-PER) (Pierce, Rockford, IL, USA) supplemented with complete ethylenediaminetetraacetic acid (EDTA)-free mini protease inhibitor (Roche, Manheim, Germany) and spun at 15,000 rpm in a 4 °C microcentrifuge to remove impurities. Luciferase expression was quantified by mixing 40 μL cell lysate with 100 μL of luciferase assay reagent (Promega, Madison, WI, USA) in a single well for each sample on a 96-well plate. β-galactosidase expression was quantified by adding 20 μL cell lysate with 100 μL of Gal Screen Reagent (Tropix, Bedford, MA, USA). The 96-well plates were incubated at room temperature, followed by quantification of luminescence by a microplate luminometer (Synergy HTX Multi-mode reader, BioTek, Winooski, VT, USA). Luminescence was measured in relative light units (RLU). The data were normalized for transfection efficiency (luciferase RLU/ β-galactosidase RLU) and further normalized to a percentage. The data shown represent five independent experiments of at least two separate samples, and figures are composite graphs representing data from at least three independent experiments.
Western immunoblots. Western immunoblots were performed by way of standard procedures. Mammalian cells were lysed in M-PER (Pierce, Rockford, IL, USA) supplemented with Complete mini EDTA-free protease inhibitors (Roche, Indianapolis, IN, USA) and protein concentrations were determined by Coomassie Plus Protein Assay (Pierce, Rockford, IL, USA). After separation on 10–20% Criterion gels (Bio-Rad, Hercules, CA, USA) proteins were transferred to polyvinylidene difluoride membranes. The primary antibodies used include mouse monoclonal α-FKBP52 (HI52D), rabbit α-human AR (Santa Cruz Biotechnology, Santa Cruz, CA, USA), and Glyceraldehyde-3-phosphate dehydrogenase (6C5; Biodesign International, Saco, MN, USA). Alkaline phosphatase-conjugated goat anti-mouse or anti-rabbit antibodies were used in conjunction with Immune-Star AP substrate (Bio-Rad, Hercules, CA, USA) before detection on X-ray film.
I-TASSER Homology Modeling. The template and target proteins were selected and extracted from NCBI:Pubmed (
http://0-www-ncbi-nlm-nih-gov.brum.beds.ac.uk/pubmed). The basic local alignment search tool (BLAST) was used to determine template sequence homology by specific parameters. To create the model, a sequence alignment file between the target and selected template sequence (HsFKBP51 and DrFKBP52, respectively, or HsFKBP52 and DrFKBP52, respectively) is required. To this aim, we used another online server, the multiple sequence alignment ClustalW2 (
http://www.ebi.ac.uk/Tools/msa/clustalw2/). The sequence for the selected template was obtained directly from the BLAST webpage or from the PDB website. The DrFKBP52 and DrFKBP52-A111V amino acid sequences were submitted to I-TASSER for homology modeling (
http://zhanglab.ccmb.med.umich.edu/I-TASSER/). The results were then individually assessed.