The Mammalian High Mobility Group Protein AT-Hook 2 (HMGA2): Biochemical and Biophysical Properties, and Its Association with Adipogenesis
Abstract
:1. Introduction
2. Biochemical and Biophysical Properties of HMGA2
3. HMGA2 in Adipogenesis
4. Conclusions and Perspectives
Funding
Conflicts of Interest
Abbreviations
| HMGA2 | Mammalian high-mobility-group protein AT-hook 2 |
| IDP | Intrinsically unstructured protein |
| d.p.c. | Days post-coitum |
| UTR | Untranslational region |
| GWA | Genome-wide association |
| SNP | Single nucleotide polymorphism |
| NSC | Neural stem cell |
| HSC | Hematopoietic stem cell |
| MSC | Mesenchymal stem cell |
| PPARγ | Peroxisome proliferator-activated receptor γ |
| NCP | Nucleosome core particle |
| CK2 | Casein kinase 2 |
| PDI-ELISA | Protein–DNA interaction enzyme-linked immunosorbent assay |
| HTS | High-throughput screening |
References
- Bianchi, M.E.; Beltrame, M. Upwardly mobile proteins. Workshop: The role of HMG proteins in chromatin structure, gene expression and neoplasia. EMBO Rep. 2000, 1, 109–114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Friedmann, M.; Holth, L.T.; Zoghbi, H.Y.; Reeves, R. Organization, inducible-expression and chromosome localization of the human HMG-I(Y) nonhistone protein gene. Nucleic Acids Res. 1993, 21, 4259–4267. [Google Scholar] [CrossRef] [Green Version]
- Manfioletti, G.; Giancotti, V.; Bandiera, A.; Buratti, E.; Sautiere, P.; Cary, P.; Crane-Robinson, C.; Coles, B.; Goodwin, G.H. cDNA cloning of the HMGI-C phosphoprotein, a nuclear protein associated with neoplastic and undifferentiated phenotypes. Nucleic Acids Res. 1991, 19, 6793–6797. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Benson, K.F.; Przybysz, K.; Liu, J.; Hou, Y.; Cherath, L.; Chada, K. Genomic structure and expression of the murine Hmgi-c gene. Nucleic Acids Res. 1996, 24, 4071–4077. [Google Scholar] [CrossRef] [PubMed]
- Hauke, S.; Flohr, A.M.; Rogalla, P.; Bullerdiek, J. Sequencing of intron 3 of HMGA2 uncovers the existence of a novel exon. Genes Chromosomes. Cancer 2002, 34, 17–23. [Google Scholar] [CrossRef]
- Goodwin, G.H.; Johns, E.W. Isolation and characterisation of two calf-thymus chromatin non-histone proteins with high contents of acidic and basic amino acids. Eur. J. Biochem. 1973, 40, 215–219. [Google Scholar] [CrossRef]
- Goodwin, G.H.; Sanders, C.; Johns, E.W. A new group of chromatin-associated proteins with a high content of acidic and basic amino acids. Eur. J. Biochem. 1973, 38, 14–19. [Google Scholar] [CrossRef]
- Bustin, M. Revised nomenclature for high mobility group (HMG) chromosomal proteins. Trends Biochem. Sci. 2001, 26, 152–153. [Google Scholar] [CrossRef]
- Johns, E.W. The HMG Chromosomal Proteins; Academic Press: London, UK, 1982; pp. 1–262. [Google Scholar]
- Lund, T.; Holtlund, J.; Fredriksen, M.; Laland, S.G. On the presence of two new high mobility group-like proteins in HeLa S3 cells. FEBS Lett. 1983, 152, 163–167. [Google Scholar] [CrossRef] [Green Version]
- Giancotti, V.; Berlingieri, M.T.; DiFiore, P.P.; Fusco, A.; Vecchio, G.; Crane-Robinson, C. Changes in nuclear proteins on transformation of rat epithelial thyroid cells by a murine sarcoma retrovirus. Cancer Res. 1985, 45, 6051–6057. [Google Scholar]
- Goodwin, G.H.; Cockerill, P.N.; Kellam, S.; Wright, C.A. Fractionation by high-performance liquid chromatography of the low- molecular-mass high-mobility-group (HMG) chromosomal proteins present in proliferating rat cells and an investigation of the HMG proteins present in virus transformed cells. Eur. J. Biochem. 1985, 149, 47–51. [Google Scholar] [CrossRef] [PubMed]
- Patel, U.A.; Bandiera, A.; Manfioletti, G.; Giancotti, V.; Chau, K.Y.; Crane-Robinson, C. Expression and cDNA cloning of human HMGI-C phosphoprotein. Biochem. Biophys. Res. Commun. 1994, 201, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Ashar, H.R.; Fejzo, M.S.; Tkachenko, A.; Zhou, X.; Fletcher, J.A.; Weremowicz, S.; Morton, C.C.; Chada, K. Disruption of the architectural factor HMGI-C: DNA-binding AT hook motifs fused in lipomas to distinct transcriptional regulatory domains. Cell 1995, 82, 57–65. [Google Scholar] [CrossRef] [Green Version]
- Schoenmakers, E.F.; Wanschura, S.; Mols, R.; Bullerdiek, J.; Van den, B.H.; Van de Ven, W.J. Recurrent rearrangements in the high mobility group protein gene, HMGI- C, in benign mesenchymal tumours. Nat. Genet. 1995, 10, 436–444. [Google Scholar] [CrossRef]
- Ashar, H.R.; Cherath, L.; Przybysz, K.M.; Chada, K. Genomic characterization of human HMGIC, a member of the accessory transcription factor family found at translocation breakpoints in lipomas. Genomics 1996, 31, 207–214. [Google Scholar] [CrossRef]
- Lee, Y.S.; Dutta, A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007, 21, 1025–1030. [Google Scholar] [CrossRef] [Green Version]
- Yu, F.; Yao, H.; Zhu, P.; Zhang, X.; Pan, Q.; Gong, C.; Huang, Y.; Hu, X.; Su, F.; Lieberman, J.; et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell 2007, 131, 1109–1123. [Google Scholar] [CrossRef] [Green Version]
- Park, S.M.; Shell, S.; Radjabi, A.R.; Schickel, R.; Feig, C.; Boyerinas, B.; Dinulescu, D.M.; Lengyel, E.; Peter, M.E. Let-7 prevents early cancer progression by suppressing expression of the embryonic gene HMGA2. Cell Cycle 2007, 6, 2585–2590. [Google Scholar] [CrossRef] [Green Version]
- Frost, L.; Baez, M.A.; Harrilal, C.; Garabedian, A.; Fernandez-Lima, F.; Leng, F. The Dimerization State of the Mammalian High Mobility Group Protein AT-Hook 2 (HMGA2). PLoS ONE 2015, 10, e0130478. [Google Scholar] [CrossRef]
- Giancotti, V.; Pani, B.; D’Andrea, P.; Berlingieri, M.T.; Di Fiore, P.P.; Fusco, A.; Vecchio, G.; Philp, R.; Crane-Robinson, C.; Nicolas, R.H. Elevated levels of a specific class of nuclear phosphoproteins in cells transformed with v-ras; v-mos oncogenes; by cotransfection with c-myc and polyoma middle T genes. EMBO J. 1987, 6, 1981–1987. [Google Scholar] [CrossRef]
- Yie, J.; Merika, M.; Munshi, N.; Chen, G.; Thanos, D. The role of HMG I(Y) in the assembly and function of the IFN-beta enhanceosome. EMBO J. 1999, 18, 3074–3089. [Google Scholar] [CrossRef] [Green Version]
- Edwards, L.K. Biochemical characterization of mammalian high mobility group protein A2. Master of Science Thesis, Florida International University, Miami, FL, USA, March 2006. [Google Scholar]
- Danielsson, J.; Liljedahl, L.; Barany-Wallje, E.; Sonderby, P.; Kristensen, L.H.; Martinez-Yamout, M.A.; Dyson, H.J.; Wright, P.E.; Poulsen, F.M.; Maler, L.; et al. The intrinsically disordered RNR inhibitor Sml1 is a dynamic dimer. Biochemistry 2008, 47, 13428–13437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sigalov, A.B. Structural biology of intrinsically disordered proteins: Revisiting unsolved mysteries. Biochimie 2016, 125, 112–118. [Google Scholar] [CrossRef] [PubMed]
- Sigalov, A.; Aivazian, D.; Stern, L. Homooligomerization of the cytoplasmic domain of the T cell receptor zeta chain and of other proteins containing the immunoreceptor tyrosine-based activation motif. Biochemistry 2004, 43, 2049–2061. [Google Scholar] [CrossRef] [PubMed]
- Lanza, D.C.; Silva, J.C.; Assmann, E.M.; Quaresma, A.J.; Bressan, G.C.; Torriani, I.L.; Kobarg, J. Human FEZ1 has characteristics of a natively unfolded protein and dimerizes in solution. Proteins 2009, 74, 104–121. [Google Scholar] [CrossRef] [PubMed]
- Simon, S.M.; Sousa, F.J.; Mohana-Borges, R.; Walker, G.C. Regulation of Escherichia coli SOS mutagenesis by dimeric intrinsically disordered umuD gene products. Proc. Natl. Acad. Sci. USA 2008, 105, 1152–1157. [Google Scholar] [CrossRef] [Green Version]
- Pieprzyk, J.; Zbela, A.; Jakob, M.; Ozyhar, A.; Orlowski, M. Homodimerization propensity of the intrinsically disordered N-terminal domain of Ultraspiracle from Aedes aegypti. Biochim. Biophys. Acta. 2014, 1844, 1153–1166. [Google Scholar] [CrossRef]
- Reeves, R.; Nissen, M.S. The AT-DNA-binding domain of mammalian high mobility group I chromosomal proteins. A novel peptide motif for recognizing DNA structure. J. Biol. Chem. 1990, 265, 8573–8582. [Google Scholar]
- Geierstanger, B.H.; Volkman, B.F.; Kremer, W.; Wemmer, D.E. Short peptide fragments derived from HMG-I/Y proteins bind specifically to the minor groove of DNA. Biochemistry 1994, 33, 5347–5355. [Google Scholar] [CrossRef]
- Huth, J.R.; Bewley, C.A.; Nissen, M.S.; Evans, J.N.; Reeves, R.; Gronenborn, A.M.; Clore, G.M. The solution structure of an HMG-I(Y)-DNA complex defines a new architectural minor groove binding motif. Nat. Struct. Biol. 1997, 4, 657–665. [Google Scholar] [CrossRef]
- Fonfria-Subiros, E.; Acosta-Reyes, F.; Saperas, N.; Pous, J.; Subirana, J.A.; Campos, J.L. Crystal structure of a complex of DNA with one AT-hook of HMGA1. PLoS ONE 2012, 7, e37120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, B.; Young, J.; Leng, F. DNA bending by the mammalian high-mobility group protein AT hook 2. Biochemistry 2010, 49, 1590–1595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schenk, E.R.; Ridgeway, M.E.; Park, M.A.; Leng, F.; Fernandez-Lima, F. Isomerization kinetics of AT hook decapeptide solution structures. Anal. Chem. 2014, 86, 1210–1214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, J.; Rauscher, S.; Nawrocki, G.; Ran, T.; Feig, M.; de Groot, B.L.; Grubmuller, H.; MacKerell, A.D., Jr. CHARMM36m: An improved force field for folded and intrinsically disordered proteins. Nat. Methods 2017, 14, 71–73. [Google Scholar] [CrossRef] [Green Version]
- Ho, B.K.; Dill, K.A. Folding very short peptides using molecular dynamics. PLoS Comput. Biol. 2006, 2, e27. [Google Scholar] [CrossRef] [Green Version]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kale, L.; Schulten, K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef] [Green Version]
- Solomon, M.J.; Strauss, F.; Varshavsky, A. A mammalian high mobility group protein recognizes any stretch of six A.T base pairs in duplex DNA. Proc. Natl. Acad. Sci. USA 1986, 83, 1276–1280. [Google Scholar] [CrossRef] [Green Version]
- Maher, J.F.; Nathans, D. Multivalent DNA-binding properties of the HMG-1 proteins. Proc. Natl. Acad. Sci. USA 1996, 93, 6716–6720. [Google Scholar] [CrossRef] [Green Version]
- Thanos, D.; Maniatis, T. The high mobility group protein HMG I(Y) is required for NF-kappa B-dependent virus induction of the human IFN-beta gene. Cell 1992, 71, 777–789. [Google Scholar] [CrossRef]
- Du, W.; Thanos, D.; Maniatis, T. Mechanisms of transcriptional synergism between distinct virus-inducible enhancer elements. Cell 1993, 74, 887–898. [Google Scholar] [CrossRef]
- Baldassarre, G.; Fedele, M.; Battista, S.; Vecchione, A.; Klein-Szanto, A.J.; Santoro, M.; Waldmann, T.A.; Azimi, N.; Croce, C.M.; Fusco, A. Onset of natural killer cell lymphomas in transgenic mice carrying a truncated HMGI-C gene by the chronic stimulation of the IL-2 and IL-15 pathway. Proc. Natl. Acad. Sci. USA 2001, 98, 7970–7975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- John, S.; Reeves, R.B.; Lin, J.X.; Child, R.; Leiden, J.M.; Thompson, C.B.; Leonard, W.J. Regulation of cell-type-specific interleukin-2 receptor alpha-chain gene expression: Potential role of physical interactions between Elf-1, HMG-I(Y), and NF-kappa B family proteins. Mol. Cell Biol. 1995, 15, 1786–1796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, T.; Leng, F. Specific recognition of AT-rich DNA sequences by the mammalian high mobility group protein AT-hook 2: A SELEX study. Biochemistry 2007, 46, 13059–13066. [Google Scholar] [CrossRef] [PubMed]
- Winter, N.; Nimzyk, R.; Bosche, C.; Meyer, A.; Bullerdiek, J. Chromatin immunoprecipitation to analyze DNA binding sites of HMGA2. PLoS ONE 2011, 6, e18837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwanbeck, R.; Manfioletti, G.; Wisniewski, J.R. Architecture of high mobility group protein I-C.DNA complex and its perturbation upon phosphorylation by Cdc2 kinase. J. Biol. Chem. 2000, 275, 1793–1801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piekielko, A.; Drung, A.; Rogalla, P.; Schwanbeck, R.; Heyduk, T.; Gerharz, M.; Bullerdiek, J.; Wisniewski, J.R. Distinct organization of DNA complexes of various HMGI/Y family proteins and their modulation upon mitotic phosphorylation. J. Biol. Chem. 2001, 276, 1984–1992. [Google Scholar] [CrossRef] [Green Version]
- Cui, T.; Wei, S.; Brew, K.; Leng, F. Energetics of binding the mammalian high mobility group protein HMGA2 to poly(dA-dT)2 and poly(dA)-poly(dT). J. Mol. Biol. 2005, 352, 629–645. [Google Scholar] [CrossRef]
- Nissen, M.S.; Reeves, R. Changes in superhelicity are introduced into closed circular DNA by binding of high mobility group protein I/Y. J. Biol. Chem. 1995, 270, 4355–4360. [Google Scholar] [CrossRef] [Green Version]
- Schwanbeck, R.; Wisniewski, J.R. Cdc2 and mitogen-activated protein kinases modulate DNA binding properties of the putative transcriptional regulator Chironomus high mobility group protein I. J. Biol. Chem. 1997, 272, 27476–27483. [Google Scholar] [CrossRef] [Green Version]
- Noro, B.; Licheri, B.; Sgarra, R.; Rustighi, A.; Tessari, M.A.; Chau, K.Y.; Ono, S.J.; Giancotti, V.; Manfioletti, G. Molecular dissection of the architectural transcription factor HMGA2. Biochemistry 2003, 42, 4569–4577. [Google Scholar] [CrossRef]
- Sgarra, R.; Zammitti, S.; Lo, S.A.; Maurizio, E.; Arnoldo, L.; Pegoraro, S.; Giancotti, V.; Manfioletti, G. HMGA molecular network: From transcriptional regulation to chromatin remodeling. Biochim. Biophys. Acta. 2009. [Google Scholar] [CrossRef] [PubMed]
- Maurizio, E.; Cravello, L.; Brady, L.; Spolaore, B.; Arnoldo, L.; Giancotti, V.; Manfioletti, G.; Sgarra, R. Conformational role for the C-terminal tail of the intrinsically disordered high mobility group A (HMGA) chromatin factors. J. Proteome. Res. 2011, 10, 3283–3291. [Google Scholar] [CrossRef] [PubMed]
- Battista, S.; Fidanza, V.; Fedele, M.; Klein-Szanto, A.J.; Outwater, E.; Brunner, H.; Santoro, M.; Croce, C.M.; Fusco, A. The expression of a truncated HMGI-C gene induces gigantism associated with lipomatosis. Cancer Res. 1999, 59, 4793–4797. [Google Scholar] [PubMed]
- Xiang, X.; Benson, K.F.; Chada, K. Mini-mouse: Disruption of the pygmy locus in a transgenic insertional mutant. Science 1990, 247, 967–969. [Google Scholar] [CrossRef] [PubMed]
- Benson, K.F.; Chada, K. Mini-mouse: Phenotypic characterization of a transgenic insertional mutant allelic to pygmy. Genet. Res. 1994, 64, 27–33. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.; Benson, K.F.; Ashar, H.R.; Chada, K. Mutation responsible for the mouse pygmy phenotype in the developmentally regulated factor HMGI-C. Nature 1995, 376, 771–774. [Google Scholar] [CrossRef]
- Ashar, H.R.; Chouinard, R.A., Jr.; Dokur, M.; Chada, K. In vivo modulation of HMGA2 expression. Biochim. Biophys. Acta (BBA) Bioenerg. 2010, 1799, 55–61. [Google Scholar]
- Anand, A.; Chada, K. In vivo modulation of Hmgic reduces obesity. Nat. Genet. 2000, 24, 377–380. [Google Scholar] [CrossRef]
- Federico, A.; Forzati, F.; Esposito, F.; Arra, C.; Palma, G.; Barbieri, A.; Palmieri, D.; Fedele, M.; Pierantoni, G.M.; De, M.I.; et al. Hmga1/Hmga2 double knock-out mice display a "superpygmy" phenotype. Biol. Open. 2014, 3, 372–378. [Google Scholar] [CrossRef] [Green Version]
- Zaidi, M.R.; Okada, Y.; Chada, K.K. Misexpression of full-length HMGA2 induces benign mesenchymal tumors in mice. Cancer Res. 2006, 66, 7453–7459. [Google Scholar] [CrossRef] [Green Version]
- Arlotta, P.; Tai, A.K.; Manfioletti, G.; Clifford, C.; Jay, G.; Ono, S.J. Transgenic mice expressing a truncated form of the high mobility group I-C protein develop adiposity and an abnormally high prevalence of lipomas. J. Biol. Chem. 2000, 275, 14394–14400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Broberg, K.; Zhang, M.; Strombeck, B.; Isaksson, M.; Nilsson, M.; Mertens, F.; Mandahl, N.; Panagopoulos, I. Fusion of RDC1 with HMGA2 in lipomas as the result of chromosome aberrations involving 2q35-37 and 12q13-15. Int. J. Oncol. 2002, 21, 321–326. [Google Scholar] [CrossRef] [PubMed]
- Ashar, H.R.; Tkachenko, A.; Shah, P.; Chada, K. HMGA2 is expressed in an allele-specific manner in human lipomas. Cancer Genet. Cytogenet. 2003, 143, 160–168. [Google Scholar] [CrossRef]
- Ligon, A.H.; Moore, S.D.; Parisi, M.A.; Mealiffe, M.E.; Harris, D.J.; Ferguson, H.L.; Quade, B.J.; Morton, C.C. Constitutional rearrangement of the architectural factor HMGA2: A novel human phenotype including overgrowth and lipomas. Am. J. Hum. Genet. 2005, 76, 340–348. [Google Scholar] [CrossRef] [Green Version]
- Weedon, M.N.; Lettre, G.; Freathy, R.M.; Lindgren, C.M.; Voight, B.F.; Perry, J.R.; Elliott, K.S.; Hackett, R.; Guiducci, C.; Shields, B.; et al. A common variant of HMGA2 is associated with adult and childhood height in the general population. Nat. Genet. 2007, 39, 1245–1250. [Google Scholar] [CrossRef] [Green Version]
- Sanna, S.; Jackson, A.U.; Nagaraja, R.; Willer, C.J.; Chen, W.M.; Bonnycastle, L.L.; Shen, H.; Timpson, N.; Lettre, G.; Usala, G.; et al. Common variants in the GDF5-UQCC region are associated with variation in human height. Nat. Genet. 2008, 40, 198–203. [Google Scholar] [CrossRef] [Green Version]
- Weedon, M.N.; Lango, H.; Lindgren, C.M.; Wallace, C.; Evans, D.M.; Mangino, M.; Freathy, R.M.; Perry, J.R.; Stevens, S.; Hall, A.S. Genome-wide association analysis identifies 20 loci that influence adult height. Nat. Genet. 2008, 40, 575–583. [Google Scholar] [CrossRef] [Green Version]
- Yang, T.L.; Guo, Y.; Zhang, L.S.; Tian, Q.; Yan, H.; Guo, Y.F.; Deng, H.W. HMGA2 is confirmed to be associated with human adult height. Ann. Hum. Genet. 2010, 74, 11–16. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.Z.; Medland, S.E.; Wright, M.J.; Henders, A.K.; Heath, A.C.; Madden, P.A.; Duncan, A.; Montgomery, G.W.; Martin, N.G.; McRae, A.F. Genome-wide association study of height and body mass index in Australian twin families. Twin. Res. Hum. Genet. 2010, 13, 179–193. [Google Scholar] [CrossRef]
- Takeshita, H.; Fujihara, J.; Soejima, M.; Koda, Y.; Kimura-Kataoka, K.; Ono, R.; Yuasa, I.; Iida, R.; Ueki, M.; Nagao, M.; et al. Confirmation that SNPs in the high mobility group-A2 gene (HMGA2) are associated with adult height in the Japanese population; wide-ranging population survey of height-related SNPs in HMGA2. Electrophoresis 2011, 32, 1844–1851. [Google Scholar] [CrossRef]
- Carty, C.L.; Johnson, N.A.; Hutter, C.M.; Reiner, A.P.; Peters, U.; Tang, H.; Kooperberg, C. Genome-wide association study of body height in African Americans: The Women’s Health Initiative SNP Health Association Resource (SHARe). Hum. Mol. Genet. 2012, 21, 711–720. [Google Scholar] [CrossRef] [Green Version]
- Horikoshi, M.; Yaghootkar, H.; Mook-Kanamori, D.O.; Sovio, U.; Taal, H.R.; Hennig, B.J.; Bradfield, J.P.; St, P.B.; Evans, D.M.; Charoen, P.; et al. New loci associated with birth weight identify genetic links between intrauterine growth and adult height and metabolism. Nat. Genet. 2013, 45, 76–82. [Google Scholar] [CrossRef] [Green Version]
- Lango, A.H.; Estrada, K.; Lettre, G.; Berndt, S.I.; Weedon, M.N.; Rivadeneira, F.; Willer, C.J.; Jackson, A.U.; Vedantam, S.; Raychaudhuri, S.; et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature 2010, 467, 832–838. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stein, J.L.; Medland, S.E.; Vasquez, A.A.; Hibar, D.P.; Senstad, R.E.; Winkler, A.M.; Toro, R.; Appel, K.; Bartecek, R.; Bergmann, O.; et al. Identification of common variants associated with human hippocampal and intracranial volumes. Nat. Genet. 2012, 44, 552–561. [Google Scholar] [CrossRef] [Green Version]
- Buysse, K.; Reardon, W.; Mehta, L.; Costa, T.; Fagerstrom, C.; Kingsbury, D.J.; Anadiotis, G.; McGillivray, B.C.; Hellemans, J.; de, L.N.; et al. The 12q14 microdeletion syndrome: Additional patients and further evidence that HMGA2 is an important genetic determinant for human height. Eur. J. Med. Genet. 2009, 52, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Lynch, S.A.; Foulds, N.; Thuresson, A.C.; Collins, A.L.; Anneren, G.; Hedberg, B.O.; Delaney, C.A.; Iremonger, J.; Murray, C.M.; Crolla, J.A.; et al. The 12q14 microdeletion syndrome: Six new cases confirming the role of HMGA2 in growth. Eur. J. Hum. Genet. 2011, 19, 534–539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosen, E.D.; MacDougald, O.A. Adipocyte differentiation from the inside out. Nat. Rev. Mol. Cell Biol. 2006, 7, 885–896. [Google Scholar] [CrossRef] [PubMed]
- Sarjeant, K.; Stephens, J.M. Adipogenesis. Cold Spring Harb. Perspect. Biol. 2012, 4, a008417. [Google Scholar] [CrossRef] [Green Version]
- Tang, Q.Q.; Lane, M.D. Adipogenesis: From stem cell to adipocyte. Annu. Rev. Biochem. 2012, 81, 715–736. [Google Scholar] [CrossRef] [Green Version]
- Ghaben, A.L.; Scherer, P.E. Adipogenesis and metabolic health. Nat. Rev. Mol. Cell Biol. 2019, 20, 242–258. [Google Scholar] [CrossRef]
- Lee, J.E.; Schmidt, H.; Lai, B.; Ge, K. Transcriptional and Epigenomic Regulation of Adipogenesis. Mol. Cell Biol. 2019, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Green, H.; Meuth, M. An established pre-adipose cell line and its differentiation in culture. Cell 1974, 3, 127–133. [Google Scholar] [CrossRef]
- Yuan, Y.; Xi, Y.; Chen, J.; Zhu, P.; Kang, J.; Zou, Z.; Wang, F.; Bu, S. STAT3 stimulates adipogenic stem cell proliferation and cooperates with HMGA2 during the early stage of differentiation to promote adipogenesis. Biochem. Biophys. Res. Commun. 2017, 482, 1360–1366. [Google Scholar] [CrossRef] [PubMed]
- Ayoubi, T.A.; Jansen, E.; Meulemans, S.M.; Van de Ven, W.J. Regulation of HMGIC expression: An architectural transcription factor involved in growth control and development. Oncogene 1999, 18, 5076–5087. [Google Scholar] [CrossRef] [Green Version]
- Vernochet, C.; Milstone, D.S.; Iehle, C.; Belmonte, N.; Phillips, B.; Wdziekonski, B.; Villageois, P.; Amri, E.Z.; O’Donnell, P.E.; Mortensen, R.M.; et al. PPARgamma-dependent and PPARgamma-independent effects on the development of adipose cells from embryonic stem cells. FEBS Lett. 2002, 510, 94–98. [Google Scholar] [CrossRef] [Green Version]
- Sun, T.; Fu, M.; Bookout, A.L.; Kliewer, S.A.; Mangelsdorf, D.J. MicroRNA let-7 regulates 3T3-L1 adipogenesis. Mol. Endocrinol. 2009, 23, 925–931. [Google Scholar] [CrossRef] [Green Version]
- Alonso, N.; Guillen, R.; Chambers, J.W.; Leng, F. A rapid and sensitive high-throughput screening method to identify compounds targeting protein-nucleic acids interactions. Nucleic Acids Res. 2015, 43, e52. [Google Scholar] [CrossRef]
- Xi, Y.; Shen, W.; Ma, L.; Zhao, M.; Zheng, J.; Bu, S.; Hino, S.; Nakao, M. HMGA2 promotes adipogenesis by activating C/EBPbeta-mediated expression of PPARgamma. Biochem. Biophys. Res. Commun. 2016, 472, 617–623. [Google Scholar] [CrossRef] [Green Version]
- Price, N.L.; Holtrup, B.; Kwei, S.L.; Wabitsch, M.; Rodeheffer, M.; Bianchini, L.; Suarez, Y.; Fernandez-Hernando, C. SREBP-1c/MicroRNA 33b Genomic Loci Control Adipocyte Differentiation. Mol. Cell Biol. 2016, 36, 1180–1193. [Google Scholar] [CrossRef] [Green Version]
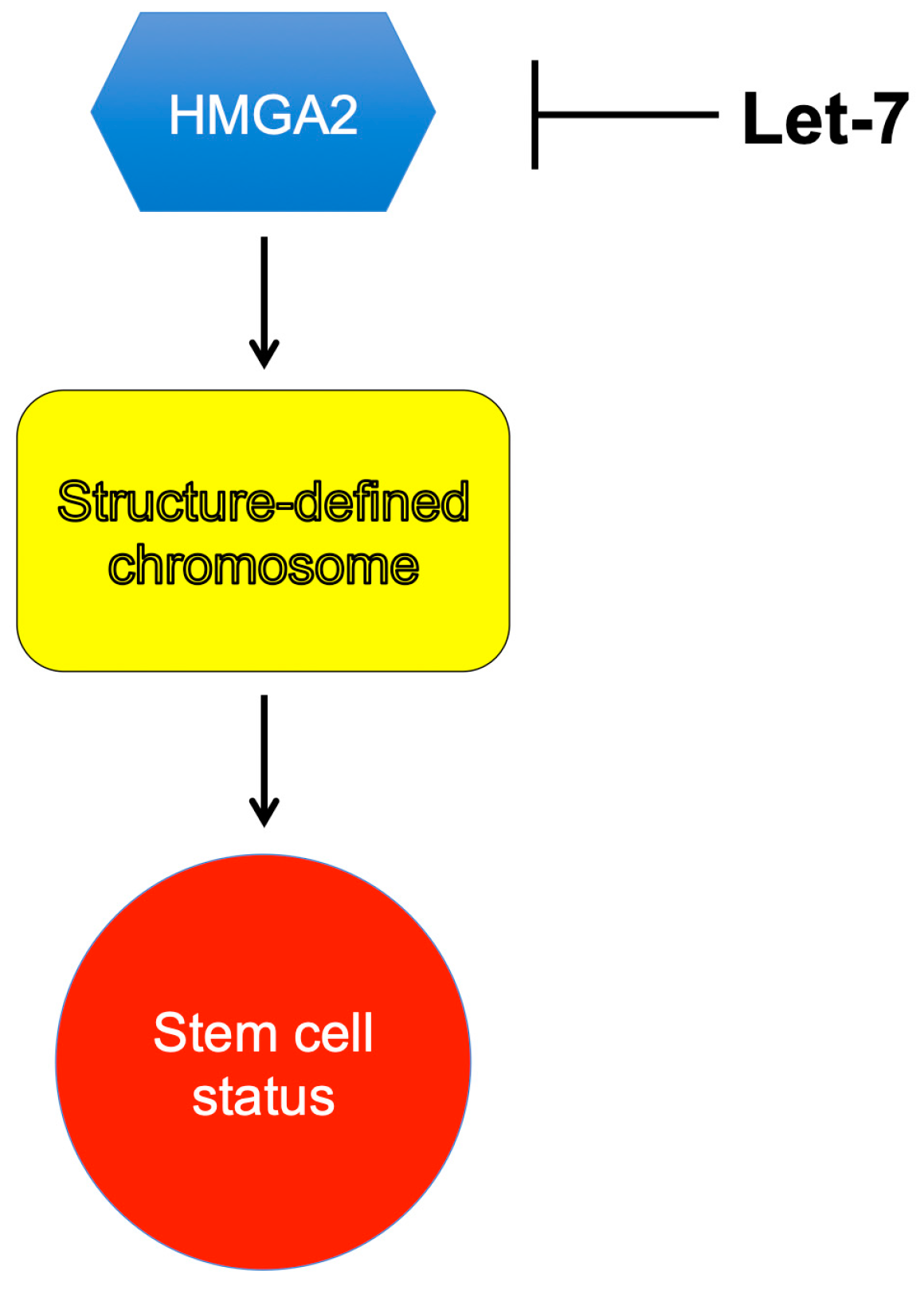
- Droge, P.; Davey, C.A. Do cells let-7 determine stemness? Cell Stem Cell 2008, 2, 8–9. [Google Scholar] [CrossRef] [Green Version]
- Hammond, S.M.; Sharpless, N.E. HMGA2, microRNAs, and stem cell aging. Cell 2008, 135, 1013–1016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pietras, E.M.; Passegue, E. Linking HSCs to their youth. Nat. Cell Biol. 2013, 15, 885–887. [Google Scholar] [CrossRef] [PubMed]
- Parisi, S.; Piscitelli, S.; Passaro, F.; Russo, T. HMGA Proteins in Stemness and Differentiation of Embryonic and Adult Stem Cells. Int. J. Mol. Sci. 2020, 21, 362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rogalla, P.; Drechsler, K.; Frey, G.; Hennig, Y.; Helmke, B.; Bonk, U.; Bullerdiek, J. HMGI-C expression patterns in human tissues. Implications for the genesis of frequent mesenchymal tumors. Am. J. Pathol. 1996, 149, 775–779. [Google Scholar]
- Gattas, G.J.; Quade, B.J.; Nowak, R.A.; Morton, C.C. HMGIC expression in human adult and fetal tissues and in uterine leiomyomata. Genes Chromosomes. Cancer 1999, 25, 316–322. [Google Scholar] [CrossRef]
- Rommel, B.; Rogalla, P.; Jox, A.; Kalle, C.V.; Kazmierczak, B.; Wolf, J.; Bullerdiek, J. HMGI-C, a member of the high mobility group family of proteins, is expressed in hematopoietic stem cells and in leukemic cells. Leuk. Lymphoma 1997, 26, 603–607. [Google Scholar] [CrossRef]
- Li, O.; Vasudevan, D.; Davey, C.A.; Droge, P. High-level expression of DNA architectural factor HMGA2 and its association with nucleosomes in human embryonic stem cells. Genesis 2006, 44, 523–529. [Google Scholar] [CrossRef]
- Nishino, J.; Kim, I.; Chada, K.; Morrison, S.J. Hmga2 promotes neural stem cell self-renewal in young but not old mice by reducing p16Ink4a and p19Arf Expression. Cell 2008, 135, 227–239. [Google Scholar] [CrossRef] [Green Version]
- Caron, L.; Bost, F.; Prot, M.; Hofman, P.; Binetruy, B. A new role for the oncogenic high-mobility group A2 transcription factor in myogenesis of embryonic stem cells. Oncogene 2005, 24, 6281–6291. [Google Scholar] [CrossRef] [Green Version]
- Markowski, D.N.; Helmke, B.M.; Meyer, F.; von, A.I.; Nimzyk, R.; Nolte, I.; Bullerdiek, J. BMP4 increases expression of HMGA2 in mesenchymal stem cells. Cytokine 2011, 56, 811–816. [Google Scholar] [CrossRef]
- Copley, M.R.; Babovic, S.; Benz, C.; Knapp, D.J.; Beer, P.A.; Kent, D.G.; Wohrer, S.; Treloar, D.Q.; Day, C.; Rowe, K.; et al. The Lin28b-let-7-Hmga2 axis determines the higher self-renewal potential of fetal haematopoietic stem cells. Nat. Cell Biol. 2013, 15, 916–925. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Li, H.; Wang, S.; Li, T.; Fan, J.; Liang, X.; Li, J.; Han, Q.; Zhu, L.; Fan, L.; et al. let-7 enhances osteogenesis and bone formation while repressing adipogenesis of human stromal/mesenchymal stem cells by regulating HMGA2. Stem Cells Dev. 2014, 23, 1452–1463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, I.; Mehta, A.; Contreras, A.; Boettger, T.; Carraro, G.; Wheeler, M.; Cabrera-Fuentes, H.A.; Bellusci, S.; Seeger, W.; Braun, T.; et al. Hmga2 is required for canonical WNT signaling during lung development. BMC. Biol. 2014, 12, 21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parameswaran, S.; Xia, X.; Hegde, G.; Ahmad, I. Hmga2 regulates self-renewal of retinal progenitors. Development 2014, 141, 4087–4097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, K.R.; Shin, J.H.; Kim, J.J.; Koog, M.G.; Lee, J.Y.; Choi, S.W.; Kim, H.S.; Seo, Y.; Lee, S.; Shin, T.H.; et al. Rapid and Efficient Direct Conversion of Human Adult Somatic Cells into Neural Stem Cells by HMGA2/let-7b. Cell Rep. 2015, 10, 441–452. [Google Scholar] [CrossRef]
- Kalomoiris, S.; Cicchetto, A.C.; Lakatos, K.; Nolta, J.A.; Fierro, F.A. Fibroblast Growth Factor 2 Regulates High Mobility Group A2 Expression in Human Bone Marrow-Derived Mesenchymal Stem Cells. J. Cell Biochem. 2016, 117, 2128–2137. [Google Scholar] [CrossRef] [Green Version]
- Rowe, R.G.; Wang, L.D.; Coma, S.; Han, A.; Mathieu, R.; Pearson, D.S.; Ross, S.; Sousa, P.; Nguyen, P.T.; Rodriguez, A.; et al. Developmental regulation of myeloerythroid progenitor function by the Lin28b-let-7-Hmga2 axis. J. Exp. Med. 2016, 213, 1497–1512. [Google Scholar] [CrossRef]
- Yu, K.R.; Park, S.B.; Jung, J.W.; Seo, M.S.; Hong, I.S.; Kim, H.S.; Seo, Y.; Kang, T.W.; Lee, J.Y.; Kurtz, A.; et al. HMGA2 regulates the in vitro aging and proliferation of human umbilical cord blood-derived stromal cells through the mTOR/p70S6K signaling pathway. Stem Cell Res. 2013, 10, 156–165. [Google Scholar] [CrossRef] [Green Version]
- Cavazzana-Calvo, M.; Payen, E.; Negre, O.; Wang, G.; Hehir, K.; Fusil, F.; Down, J.; Denaro, M.; Brady, T.; Westerman, K.; et al. Transfusion independence and HMGA2 activation after gene therapy of human beta-thalassaemia. Nature 2010, 467, 318–322. [Google Scholar] [CrossRef]
- Zhou, X.; Chada, K. HMGI family proteins: Architectural transcription factors in mammalian development and cancer. Keio J. Med. 1998, 47, 73–77. [Google Scholar] [CrossRef]
- Tallini, G.; Dal Cin, P. HMGI(Y) and HMGI-C dysregulation: A common occurrence in human tumors. Adv. Anat. Pathol. 1999, 6, 237–246. [Google Scholar] [CrossRef]
- Reeves, R.; Beckerbauer, L.M. HMGA proteins as therapeutic drug targets. Prog. Cell Cycle Res. 2003, 5, 279–286. [Google Scholar] [PubMed]
- Reeves, R. HMGA proteins: Flexibility finds a nuclear niche? Biochem. Cell Biol. 2003, 81, 185–195. [Google Scholar] [CrossRef] [PubMed]
- Young, A.R.; Narita, M. Oncogenic HMGA2: Short or small? Genes Dev. 2007, 21, 1005–1009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fusco, A.; Fedele, M. Roles of HMGA proteins in cancer. Nat. Rev. Cancer 2007, 7, 899–910. [Google Scholar] [CrossRef]
- Cleynen, I.; Van de Ven, W.J. The HMGA proteins: A myriad of functions (Review). Int. J. Oncol. 2008, 32, 289–305. [Google Scholar] [CrossRef] [Green Version]
- Fedele, M.; Fusco, A. HMGA and cancer. Biochim. Biophys. Acta. 2010, 1799, 48–54. [Google Scholar] [CrossRef]
- Sgarra, R.; Pegoraro, S.; Ros, G.; Penzo, C.; Chiefari, E.; Foti, D.; Brunetti, A.; Manfioletti, G. High Mobility Group A (HMGA) proteins: Molecular instigators of breast cancer onset and progression. Biochim. Biophys. Acta Rev. Cancer 2018, 1869, 216–229. [Google Scholar] [CrossRef]
- Zhang, S.; Mo, Q.; Wang, X. Oncological role of HMGA2 (Review). Int. J. Oncol. 2019, 55, 775–788. [Google Scholar] [CrossRef]
- Ozturk, N.; Singh, I.; Mehta, A.; Braun, T.; Barreto, G. HMGA proteins as modulators of chromatin structure during transcriptional activation. Front. Cell Dev. Biol. 2014, 2, 5. [Google Scholar] [CrossRef] [Green Version]
- Giancotti, V.; Bergamin, N.; Cataldi, P.; Rizzi, C. Epigenetic Contribution of High-Mobility Group A Proteins to Stem Cell Properties. Int. J. Cell Biol. 2018, 2018, 3698078. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, H.; Lim, H.H.; Tjokro, N.O.; Sathiyanathan, P.; Natarajan, S.; Chew, T.W.; Klonisch, T.; Goodman, S.D.; Surana, U.; Droge, P. Chaperoning HMGA2 protein protects stalled replication forks in stem and cancer cells. Cell Rep. 2014, 6, 684–697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmed, S.M.; Ramani, P.D.; Wong, S.Q.R.; Zhao, X.; Ivanyi-Nagy, R.; Leong, T.C.; Chua, C.; Li, Z.; Hentze, H.; Tan, I.B.; et al. The chromatin structuring protein HMGA2 influences human subtelomere stability and cancer chemosensitivity. PLoS ONE 2019, 14, e0215696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmed, S.M.; Droge, P. Oncofetal HMGA2 attenuates genotoxic damage induced by topoisomerase II target compounds through the regulation of local DNA topology. Mol. Oncol. 2019. [Google Scholar] [CrossRef] [Green Version]
- Goodwin, G.H.; Mathew, C.G.; Wright, C.A.; Venkov, C.D.; Johns, E.W. Analysis of the high mobility group proteins associated with salt- soluble nucleosomes. Nucleic Acids Res. 1979, 7, 1815–1835. [Google Scholar] [CrossRef] [Green Version]
- Cao, X.; Clavijo, C.; Li, X.; Lin, H.H.; Chen, Y.; Shih, H.M.; Ann, D.K. SUMOylation of HMGA2: Selective destabilization of promyelocytic leukemia protein via proteasome. Mol. Cancer Ther. 2008, 7, 923–934. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Wang, X.; Xu, F.; Zhang, L.; Wang, T.; Fu, X.; Jin, T.; Zhang, W.; Ye, L. The regulation of acetylation and stability of HMGA2 via the HBXIP-activated Akt-PCAF pathway in promotion of esophageal squamous cell carcinoma growth. Nucleic Acids Res. 2020, 48, 4858–4876. [Google Scholar] [CrossRef] [Green Version]
- Bustin, M.; Reeves, R. High-mobility-group chromosomal proteins: Architectural components that facilitate chromatin function. Prog. Nucleic Acid Res. Mol. Biol. 1996, 54, 35–100. [Google Scholar]
- Rogalla, P.; Drechsler, K.; Schroder-Babo, W.; Eberhardt, K.; Bullerdiek, J. HMGIC expression patterns in non-small lung cancer and surrounding tissue. Anticancer Res. 1998, 18, 3327–3330. [Google Scholar]
- Kumar, M.S.; Armenteros-Monterroso, E.; East, P.; Chakravorty, P.; Matthews, N.; Winslow, M.M.; Downward, J. HMGA2 functions as a competing endogenous RNA to promote lung cancer progression. Nature 2014, 505, 212–217. [Google Scholar] [CrossRef]
- Sun, M.; Song, C.X.; Huang, H.; Frankenberger, C.A.; Sankarasharma, D.; Gomes, S.; Chen, P.; Chen, J.; Chada, K.K.; He, C.; et al. HMGA2/TET1/HOXA9 signaling pathway regulates breast cancer growth and metastasis. Proc. Natl. Acad. Sci. USA 2013, 110, 9920–9925. [Google Scholar] [CrossRef] [Green Version]
- El, A.I.; Fatima, I.; Wend, P.; Alva-Ornelas, J.A.; Runke, S.; Kuenzinger, W.L.; Silva, J.; Silva, W.; Gray, J.K.; Lehr, S.; et al. The WNT10B Network Is Associated with Survival and Metastases in Chemoresistant Triple-Negative Breast Cancer. Cancer Res. 2019, 79, 982–993. [Google Scholar]
- Muller, M.H.; Drieschner, N.; Focken, T.; Bartnitzke, S.; Winter, N.; Klemke, M.; Bullerdiek, J. HMGA2 expression in the PC-3 prostate cancer cell line is autonomous of growth factor stimulation. Anticancer Res. 2013, 33, 3069–3078. [Google Scholar]
- Marquis, M.; Beaubois, C.; Lavallee, V.P.; Abrahamowicz, M.; Danieli, C.; Lemieux, S.; Ahmad, I.; Wei, A.; Ting, S.B.; Fleming, S.; et al. High expression of HMGA2 independently predicts poor clinical outcomes in acute myeloid leukemia. Blood Cancer J. 2018, 8, 68. [Google Scholar] [CrossRef] [PubMed]
- Murakami, M.; Suzuki, M.; Nishino, Y.; Funaba, M. Regulatory expression of genes related to metastasis by TGF-beta and activin A in B16 murine melanoma cells. Mol. Biol. Rep. 2010, 37, 1279–1286. [Google Scholar] [CrossRef] [PubMed]
- Raskin, L.; Fullen, D.R.; Giordano, T.J.; Thomas, D.G.; Frohm, M.L.; Cha, K.B.; Ahn, J.; Mukherjee, B.; Johnson, T.M.; Gruber, S.B. Transcriptome profiling identifies HMGA2 as a biomarker of melanoma progression and prognosis. J. Investig. Dermatol. 2013, 133, 2585–2592. [Google Scholar] [CrossRef] [Green Version]
- Hou, X.; Wan, W.; Wang, J.; Li, M.; Wang, Y.; Yao, Y.; Feng, L.; Jing, L.; Lu, H.; Jia, Y.; et al. Let-7a inhibits migration of melanoma cells via down-regulation of HMGA2 expression. Am. J. Transl. Res. 2016, 8, 3656–3665. [Google Scholar]
- Moon, H.; Donahue, L.R.; Choi, E.; Scumpia, P.O.; Lowry, W.E.; Grenier, J.K.; Zhu, J.; White, A.C. Melanocyte Stem Cell Activation and Translocation Initiate Cutaneous Melanoma in Response to UV Exposure. Cell Stem Cell 2017, 21, 665–678. [Google Scholar] [CrossRef] [Green Version]
- Miao, Y.; Cui, T.; Leng, F.; Wilson, W.D. Inhibition of high-mobility-group A2 protein binding to DNA by netropsin: A biosensor-surface plasmon resonance assay. Anal. Biochem. 2008, 374, 7–15. [Google Scholar] [CrossRef] [Green Version]
- Neidle, S. DNA minor-groove recognition by small molecules. Nat. Prod. Rep. 2001, 18, 291–309. [Google Scholar] [CrossRef]
- Zimmer, C.; Wahnert, U. Nonintercalating DNA-binding ligands: Specificity of the interaction and their use as tools in biophysical, biochemical and biological investigations of the genetic material. Prog. Biophys. Mol. Biol. 1986, 47, 31–112. [Google Scholar] [CrossRef]
- Wemmer, D.E. Designed sequence-specific minor groove ligands. Annu. Rev. Biophys. Biomol. Struct. 2000, 29, 439–461. [Google Scholar] [CrossRef] [PubMed]
- Baraldi, P.G.; Bovero, A.; Fruttarolo, F.; Preti, D.; Tabrizi, M.A.; Pavani, M.G.; Romagnoli, R. DNA minor groove binders as potential antitumor and antimicrobial agents. Med. Res. Rev. 2004, 24, 475–528. [Google Scholar] [CrossRef] [PubMed]
- Dervan, P.B. Design of sequence-specific DNA-binding molecules. Science 1986, 232, 464–471. [Google Scholar] [CrossRef] [Green Version]
- Wemmer, D.E.; Dervan, P.B. Targeting the minor groove of DNA. Curr. Opin. Struct. Biol. 1997, 7, 355–361. [Google Scholar] [CrossRef]
- Dervan, P.B.; Burli, R.W. Sequence-specific DNA recognition by polyamides. Curr. Opin. Chem. Biol. 1999, 3, 688–693. [Google Scholar] [CrossRef]
- Yan, C.; Higgins, P.J. Drugging the undruggable: Transcription therapy for cancer. Biochim. Biophys. Acta. 2013, 1835, 76–85. [Google Scholar] [CrossRef] [Green Version]
- Lambert, M.; Jambon, S.; Depauw, S.; David-Cordonnier, M.H. Targeting Transcription Factors for Cancer Treatment. Molecules. 2018, 23, 1479. [Google Scholar] [CrossRef] [Green Version]
- Helmer, D.; Schmitz, K. Peptides and Peptide Analogs to Inhibit Protein-Protein Interactions. Adv. Exp. Med. Biol. 2016, 917, 147–183. [Google Scholar]
- Rai, J. Peptide and protein mimetics by retro and retroinverso analogs. Chem. Biol. Drug Des. 2019, 93, 724–736. [Google Scholar] [CrossRef]
- French, S.W.; Schmidt, M.C.; Glorioso, J.C. Involvement of a high-mobility-group protein in the transcriptional activity of herpes simplex virus latency-active promoter 2. Mol. Cell Biol. 1996, 16, 5393–5399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berlingieri, M.T.; Manfioletti, G.; Santoro, M.; Bandiera, A.; Visconti, R.; Giancotti, V.; Fusco, A. Inhibition of HMGI-C protein synthesis suppresses retrovirally induced neoplastic transformation of rat thyroid cells. Mol. Cell Biol. 1995, 15, 1545–1553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thuault, S.; Valcourt, U.; Petersen, M.; Manfioletti, G.; Heldin, C.H.; Moustakas, A. Transforming growth factor-beta employs HMGA2 to elicit epithelial-mesenchymal transition. J. Cell Biol. 2006, 174, 175–183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thuault, S.; Tan, E.J.; Peinado, H.; Cano, A.; Heldin, C.H.; Moustakas, A. HMGA2 and Smads co-regulate SNAIL1 expression during induction of epithelial-to-mesenchymal transition. J. Biol. Chem. 2008, 283, 33437–33446. [Google Scholar] [CrossRef] [PubMed] [Green Version]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Su, L.; Deng, Z.; Leng, F. The Mammalian High Mobility Group Protein AT-Hook 2 (HMGA2): Biochemical and Biophysical Properties, and Its Association with Adipogenesis. Int. J. Mol. Sci. 2020, 21, 3710. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21103710
Su L, Deng Z, Leng F. The Mammalian High Mobility Group Protein AT-Hook 2 (HMGA2): Biochemical and Biophysical Properties, and Its Association with Adipogenesis. International Journal of Molecular Sciences. 2020; 21(10):3710. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21103710
Chicago/Turabian StyleSu, Linjia, Zifang Deng, and Fenfei Leng. 2020. "The Mammalian High Mobility Group Protein AT-Hook 2 (HMGA2): Biochemical and Biophysical Properties, and Its Association with Adipogenesis" International Journal of Molecular Sciences 21, no. 10: 3710. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21103710




